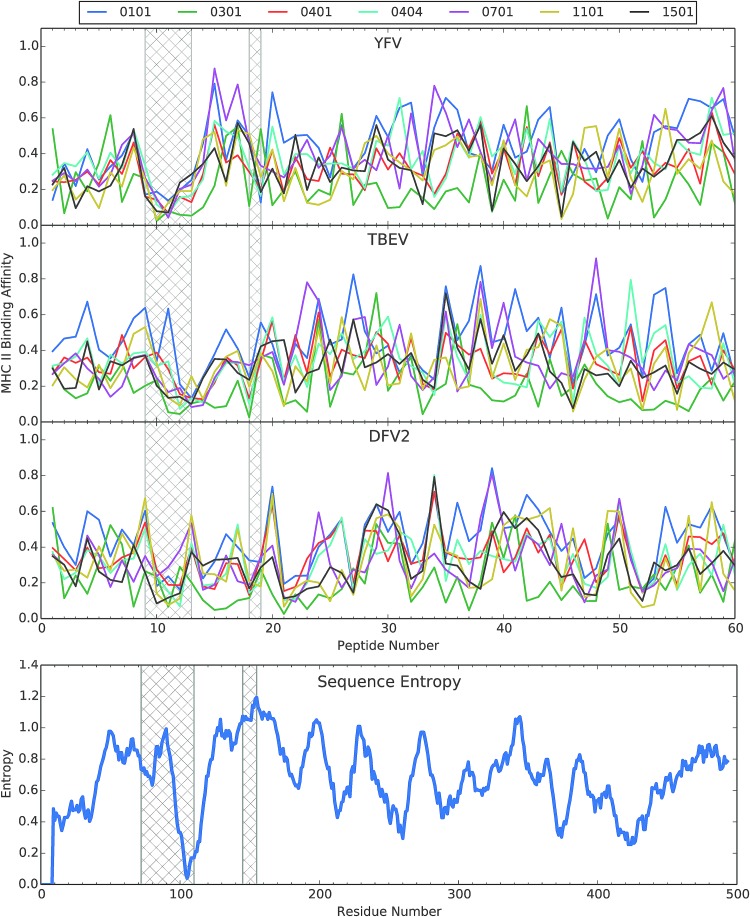
FIG. 3.
Alignment of predicted MHCII binding to selected HLA molecules of peptides from three flaviviruses and the plot of Shannon sequence entropy for an alignment of 21 flaviviruses. Hatched bars indicate the two gaps in the experimental CD4+ epitope-mapping profiles, as identified in Figure 2. MHCII binding was predicted using the NETMHCII 2.2 server (40) and then the values of “1-log50k” for each profile were normalized to the range of 0 to 1. The selected HLA molecules were those used in the tetramer-guided mapping of epitopes in the E protein for YFV (18). YFV, yellow fever virus.