ABSTRACT
We deleted subunits I (cydA) and II (cydB) of the Mycobacterium tuberculosis cytochrome bd menaquinol oxidase. The resulting ΔcydA and ΔcydAB mutants were hypersusceptible to compounds targeting the mycobacterial bc1 menaquinol-cytochrome c oxidoreductase and exhibited bioenergetic profiles indistinguishable from strains deficient in the ABC-type transporter, CydDC, predicted to be essential for cytochrome bd assembly. These results confirm CydAB and CydDC as potential targets for drugs aimed at inhibiting a terminal respiratory oxidase implicated in pathogenesis.
KEYWORDS: TB drug discovery, mycobacterial respiration, electron transport chain, extracellular flux analysis, oxidative phosphorylation
TEXT
There is resurgent interest in mycobacterial respiration and energy metabolism as potential sources of new targets and improved compounds for tuberculosis (TB) chemotherapy (1–3). This has been fueled primarily by the success of bedaquiline (BDQ), a diarylquinoline that inhibits the mycobacterial ATP synthase (4) and is approved for clinical use against multidrug-resistant (MDR) TB (5). However, additional agents in the TB drug discovery pipeline include Q203, an imidazopyridine that targets the mycobacterial cytochrome bc1 complex (6), as well as the repurposed drug, clofazimine, which acts via a redox cycling mechanism involving reduction by the type II NADH dehydrogenase followed by nonenzymatic oxidation that produces reactive oxygen species (7). Moreover, a number of recent studies have demonstrated the potential to inhibit other components of the mycobacterial electron transport chain (ETC) (1, 2, 8) as well as the opportunities inherent in simultaneously targeting multiple components of mycobacterial oxidative phosphorylation (3, 9).
The rationale is strong: respiration is essential for the survival of replicating and nonreplicating bacilli (10). In addition, while the flexibility inherent in the multiply branched mycobacterial electron transport chain implies redundancy (8), the dependence of Mycobacterium tuberculosis on a single lipoquinone, menaquinone, and only two terminal respiratory oxidases—the aa3-type cytochrome c oxidase and the cytochrome bd menaquinol oxidase (11)—suggests the potential for targeted disruption of respiratory function for both adjunctive (12) and combination (3) strategies. The cydAB-encoded cytochrome bd functions as the terminal acceptor in M. tuberculosis under microaerophilic conditions (11) and is also able to support aerobic growth during chemical inhibition of the bc1 complex, QcrCAB; for example, following exposure to imidazopyridines (3, 13).
In Escherichia coli, assembly of cytochrome bd is dependent on the ABC-type transporter, CydDC, which is also required for the synthesis of other periplasmic cytochromes (14). As a result, cydAB and cydDC mutants of E. coli exhibit overlapping, but distinct, phenotypes, consistent with the genomic separation of the two operons. In contrast, the cydDC genes in M. tuberculosis are operonic with cydAB. At the inception of this study, it was not known whether M. tuberculosis CydDC functioned solely in cytochrome bd biosynthesis; previous reports exploited a cydC::aph mutant (15), in which only the terminal gene of the cydABDC locus was eliminated, or a ΔcydA::hyg mutant (12), which was expected to disrupt full operon function owing to polar effects. There were also two articles that utilized a knockout mutant, the “cydKO” strain, reportedly lacking the 3′ end of cydB, the entire cydD, and the 5′ end of cydC (9, 13); however, a subsequent author correction to reference 13 has noted that the strain actually employed in those papers was the cydC::aph mutant (15). It was not clear, therefore, when we initiated the current study whether disruption of the entire locus was phenotypically equivalent to targeted deletion (or, by implication, chemical inhibition) of the individual genes; moreover, no reports at the time had attempted to unlink the effects of disrupted cytochrome bd menaquinol oxidase function (cydAB inactivation) from deficient ABC transport (cydDC inactivation).
In a key study published during the preparation of the manuscript, Berney, Pethe, and colleagues (3) reported that targeted disruption of cydAB eliminated oxygen respiration in M. tuberculosis bacilli exposed to Q203, killing the cells and rendering the resulting ΔcydAB mutant strain hypersusceptible to Q203 treatment in vitro in both replicating and nonreplicating (tolerant) conditions as well as in a mouse model. In that case, ΔcydAB mutants were constructed using a phage-mediated unmarking (16) system that leaves an approximately 130-bp “scar” at the deletion site following activity of the γδ resolvase.
Here, we generated targeted, in-frame deletion mutants of cydA and cydAB in M. tuberculosis H37RvMA (17) using two-step allelic exchange mutagenesis (18) in order to preserve the sequence integrity of the locus. This was confirmed by PCR and whole-genome sequencing (see Table S1 in the supplemental material). In standard microplate-based alamarBlue assays (MABA) (19) using Middlebrook 7H9 liquid growth medium (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.5% glycerol, and 0.05% Tween 80, the MIC90 values recorded for a representative panel of approved anti-TB agents from different antibiotic classes and with diverse mechanisms of action were the same for cydA and cydAB mutants and identical to those observed for wild-type M. tuberculosis H37Rv and the cydKO (cydC::aph) mutant strain (Table 1). That is, no hypersensitivity phenotype was observed for any of the cyd mutants against any of the agents tested. This included BDQ, which was in contrast to some reports (12) but consistent with other recent results (3, 9). All three cyd mutants were, however, hypersusceptible to experimental compounds for which resistance maps to qcrB (Table 1; see also Fig. S1 in the supplemental material). Moreover, there were no significant differences in MIC values across the cydA, cydAB, and cydKO (cydC::aph) strains, suggesting that elimination of either the CydAB oxidase or CydDC transport subunits was sufficient to abrogate cytochrome bd function.
TABLE 1.
MIC determinations against wild-type M. tuberculosis H37Rv and cyd mutant strains
| Compound |
M. tuberculosis straina |
|||
|---|---|---|---|---|
| H37Rv | cydKO (cydC::aph) mutant strain | ΔcydA mutant strain | ΔcydAB mutant strain | |
| Rifampin | 0.01 | 0.01 | 0.01 | 0.01 |
| Isoniazid | 0.04 | 0.04 | 0.04 | 0.04 |
| Streptomycin | 0.9 | 0.9 | 0.9 | 0.9 |
| Ethambutol | 0.47 | 0.47 | 0.47 | 0.47 |
| Pretomanid (PA-824) | 0.1 | 0.1 | 0.1 | 0.1 |
| Levofloxacin | 0.94 | 0.94 | 0.94 | 0.94 |
| BDQ | 0.03 | 0.03–0.06 | 0.03 | 0.03–0.06 |
| Q203 | 0.0097 (>50)c | 0.0003 | 0.0012 | 0.0003 |
| Compound 1b | 3.125 (>50)c | 0.39 | 0.39 | 0.39 |
| Compound 2b | 0.390 (>25)c | 0.02 | 0.0488 | 0.02 |
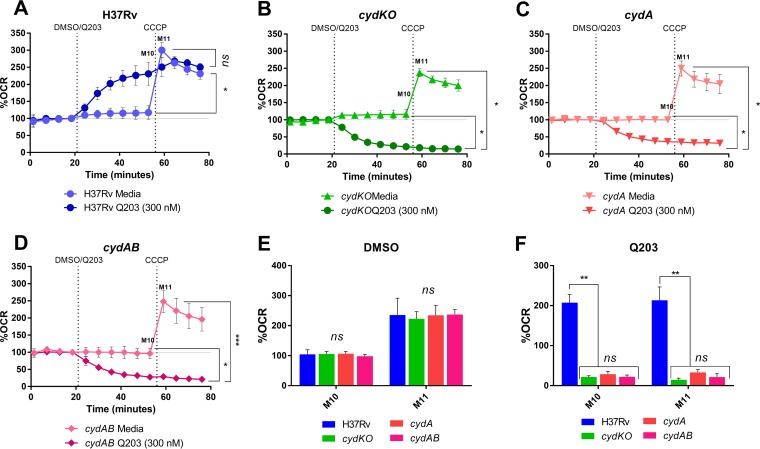
To investigate the impact of the different cyd alleles on mycobacterial respiratory function, we determined the bioenergetic responses of the cydKO (cydC::aph), cydA, and cydAB strains to Q203 treatment. Previously, we showed that Q203 inhibits electron flux through the M. tuberculosis cytochrome bc1, and this block is alleviated by rerouting electrons through cytochrome bd (9). Moreover, to compensate for the decreased proton motive force (PMF) generated by cytochrome bd and consequent drop in ATP production, bacilli increase the total electron flux through the alternative terminal oxidase, with a subsequent increase in oxygen consumption rate (OCR) (9). In contrast, in the cydKO mutant strain, Q203 treatment is associated with a rapid decrease in OCR owing to the complete inhibition of terminal oxidase function.
As observed previously (9), exposure of wild-type M. tuberculosis H37Rv to Q203 caused a significant increase in the OCR from basal levels (measurement ten [M10]) (Fig. 1A). In contrast, OCR decreased in the cydKO (cydC::aph) (M10 in Fig. 1B) strain, an effect which was also observed in the cydA and cydAB strains (M10 in Fig. 1C and D) and was consistent with the phenotype of the ΔcydAB mutant described by Berney, Pethe, and colleagues (3). Following addition of the protonophore, carbonyl cyanide m-chlorophenylhydrazone (CCCP), there was no increase in OCR in any of the Q203-treated cyd mutants (M11 in Fig. 1B to D); this was in contrast to that in wild-type M. tuberculosis H37Rv, as well as that in the mock (dimethyl sulfoxide [DMSO])-treated cyd mutants (M11 in Fig. 1A to D), and indicated the complete shutdown of electron flux through the ETC in the absence of functional cytochrome bd. No significant differences were detected in OCR levels (M10 and M11) of all strains—wild-type strain and cyd mutants—treated with DMSO (Fig. 1E). This indicated that, in the absence of Q203 treatment, ETC function of the cyd deletion mutants was similar to that of the wild type; basal OCR levels were the same (M10), and all strains exhibited comparable capacity to raise OCR to maintain membrane potential upon uncoupling through CCCP addition (M11). Moreover, the OCR levels of the Q203-treated cyd strains were equivalent, and all three mutants exhibited the same inability to elevate OCR levels after CCCP exposure (Fig. 1F).
FIG 1.
The cyd operon mutants are characterized by near-identical bioenergetics profiles. (A to D) The OCR profiles of wild-type M. tuberculosis H37RvMA and the cydKO (cydC::aph), ΔcydA, and ΔcydAB mutant strains treated with 300 nM Q203 and DMSO (as vehicle control), respectively. All plots are representative of three independent experiments, and the statistical analysis was by analysis of variance (ANOVA) (95% confidence interval) for the three biological replicates. ns, not significant; *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (ANOVA, GraphPad Prism 6.05).
In combination, our results indicate that disruption of any of the cyd operon genes (or any combination thereof) results in a cytochrome bd functionally deficient mutant characterized by a common, but distinct, bioenergetic profile that is consistent with the observed hypersusceptibility to compounds inhibiting the cytochrome c respiratory oxidase. As such, these observations offer support to recent work which has provided compelling evidence of the potential for pathway-specific combination therapies to cripple metabolic escape mechanisms, thereby enhancing compound cidality and eliminating drug-tolerant bacilli (3).
Supplementary Material
ACKNOWLEDGMENTS
We thank Vinayak Singh for technical assistance.
This work was supported by the Strategic Health Innovation Partnerships (SHIP) initiative of the South African Medical Research Council (to D.F.W. and A.J.C.S.), the South African Medical Research Council (to V.M.), the National Research Foundation of South Africa (to V.M.), the Intramural Research Program of the NIAID, NIH (to C.E.B.), and the Foundation for the National Institutes of Health with support from the Bill & Melinda Gates Foundation (to C.E.B. and V.M.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01338-17.
REFERENCES
- 1.Bald D, Villellas C, Lu P, Koul A. 2017. Targeting energy metabolism in Mycobacterium tuberculosis, a new paradigm in antimycobacterial drug discovery. mBio 8:e00272-17. doi: 10.1128/mBio.00272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sukheja P, Kumar P, Mittal N, Li SG, Singleton E, Russo R, Perryman AL, Shrestha R, Awasthi D, Husain S, Soteropoulos P, Brukh R, Connell N, Freundlich JS, Alland D. 2017. A novel small-molecule inhibitor of the Mycobacterium tuberculosis demethylmenaquinone methyltransferase MenG is bactericidal to both growing and nutritionally deprived persister cells. mBio 8:e02022-16. doi: 10.1128/mBio.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalia NP, Hasenoehrl EJ, Ab Rahman NB, Koh VH, Ang MLT, Sajorda DR, Hards K, Grüber G, Alonso S, Cook GM, Berney M, Pethe K. 2017. Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc Natl Acad Sci U S A 114:7426–7431. doi: 10.1073/pnas.1706139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 5.Dheda K, Gumbo T, Maartens G, Dooley KE, McNerney R, Murray M, Furin J, Nardell EA, London L, Lessem E, Theron G, van Helden P, Niemann S, Merker M, Dowdy D, Van Rie A, Siu GK, Pasipanodya JG, Rodrigues C, Clark TG, Sirgel FA, Esmail A, Lin HH, Atre SR, Schaaf HS, Chang KC, Lange C, Nahid P, Udwadia ZF, Horsburgh CR Jr, Churchyard GJ, Menzies D, Hesseling AC, Nuermberger E, McIlleron H, Fennelly KP, Goemaere E, Jaramillo E, Low M, Jara CM, Padayatchi N, Warren RM. 2017. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir Med 5:291–360. doi: 10.1016/S2213-2600(17)30079-6. [DOI] [PubMed] [Google Scholar]
- 6.Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, Jiricek J, Jung J, Jeon HK, Cechetto J, Christophe T, Lee H, Kempf M, Jackson M, Lenaerts AJ, Pham H, Jones V, Seo MJ, Kim YM, Seo M, Seo JJ, Park D, Ko Y, Choi I, Kim R, Kim SY, Lim S, Yim SA, Nam J, Kang H, Kwon H, Oh CT, Cho Y, Jang Y, Kim J, Chua A, Tan BH, Nanjundappa MB, Rao SP, Barnes WS, Wintjens R, Walker JR, Alonso S, Lee S, Kim J, Oh S, Oh T, Nehrbass U, Han SJ, No Z, et al. . 2013. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med 19:1157–1160. doi: 10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- 7.Yano T, Kassovska-Bratinova S, Teh JS, Winkler J, Sullivan K, Isaacs A, Schechter NM, Rubin H. 2011. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: a pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem 286:10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black PA, Warren RM, Louw GE, van Helden PD, Victor TC, Kana BD. 2014. Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2491–2503. doi: 10.1128/AAC.02293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamprecht DA, Finin PM, Rahman MA, Cumming BM, Russell SL, Jonnala SR, Adamson JH, Steyn AJ. 2016. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat Commun 7:12393. doi: 10.1038/ncomms12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boshoff HI, Barry CE III. 2005. Tuberculosis-metabolism and respiration in the absence of growth. Nat Rev Microbiol 3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 11.Cook GM, Hards K, Vilcheze C, Hartman T, Berney M. 2014. Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MGM2-0015-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berney M, Hartman TE, Jacobs WR Jr. 2014. A Mycobacterium tuberculosis cytochrome bd oxidase mutant is hypersensitive to bedaquiline. mBio 5:e01275-14. doi: 10.1128/mBio.01275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arora K, Ochoa-Montano B, Tsang PS, Blundell TL, Dawes SS, Mizrahi V, Bayliss T, Mackenzie CJ, Cleghorn LA, Ray PC, Wyatt PG, Uh E, Lee J, Barry CE III, Boshoff HI. 2014. Respiratory flexibility in response to inhibition of cytochrome c oxidase in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:6962–6965. doi: 10.1128/AAC.03486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook GM, Cruz-Ramos H, Moir AJ, Poole RK. 2002. A novel haem compound accumulated in Escherichia coli overexpressing the cydDC operon, encoding an ABC-type transporter required for cytochrome assembly. Arch Microbiol 178:358–369. doi: 10.1007/s00203-002-0467-6. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. 2005. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci U S A 102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain P, Hsu T, Arai M, Biermann K, Thaler DS, Nguyen A, Gonzalez PA, Tufariello JM, Kriakov J, Chen B, Larsen MH, Jacobs WR Jr. 2014. Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio 5:e01245-14. doi: 10.1128/mBio.01245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioerger TR, Feng Y, Ganesula K, Chen X, Dobos KM, Fortune S, Jacobs WR Jr, Mizrahi V, Parish T, Rubin E, Sassetti C, Sacchettini JC. 2010. Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J Bacteriol 192:3645–3653. doi: 10.1128/JB.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopinath K, Warner DF, Mizrahi V. 2015. Targeted gene knockout and essentiality testing by homologous recombination. Methods Mol Biol 1285:131–149. doi: 10.1007/978-1-4939-2450-9_8. [DOI] [PubMed] [Google Scholar]
- 19.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol 36:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.