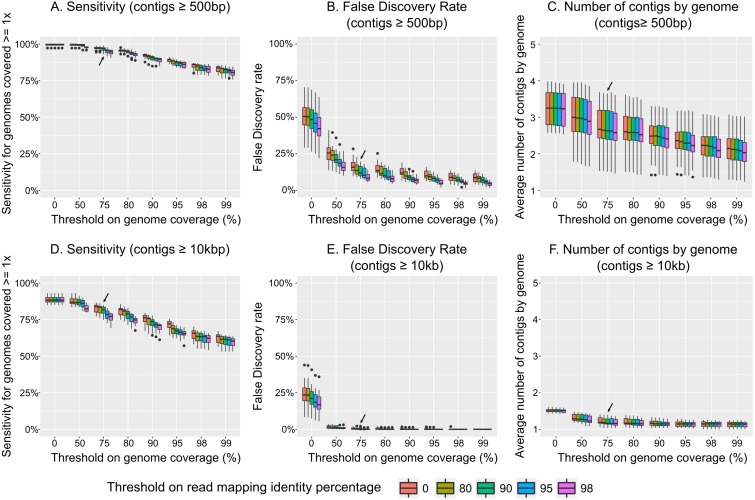
Figure 3. Impact of read mapping thresholds on accuracy of viral population detection.
Two parameters were investigated when parsing the mapping of individual virome reads to the population contigs pool: (i) the percentage of a contig covered by a sample to considered the contig as detected (x-axis), and (ii) the percentage of identity of reads mapping to the contig (color scale). Two pools of population contigs were tested: all non-redundant contigs of ≥500 bp (A–C), and all non-redundant contigs ≥10 kb (D–F). Three metrics were calculated to evaluate the impact of mapping reads thresholds. The detection sensitivity is estimated as the percentage of “expected” genomes (i.e., genomes covered ≥1 × in the sample) that were detected through mapping to population contigs (A and D). The false-discovery rate corresponds to the percentage of contigs detected in a sample through mapping to population contigs, but were not associated with any genomes from the initial sample (i.e., these genomes did not provide any reads to the simulated virome, so these contigs should not be detected, B and E). Finally the average number of distinct population contigs detected is calculated for each individual genome initially covered ≥1 ×, and correspond to the number of times a single genome is “counted” (i.e., multiple contigs suggest multiple populations, even though it is really just one population, C and F).