Fig. 4. Effects of blocking oriC-dependent replication on growth of the RNase H deficient strains.
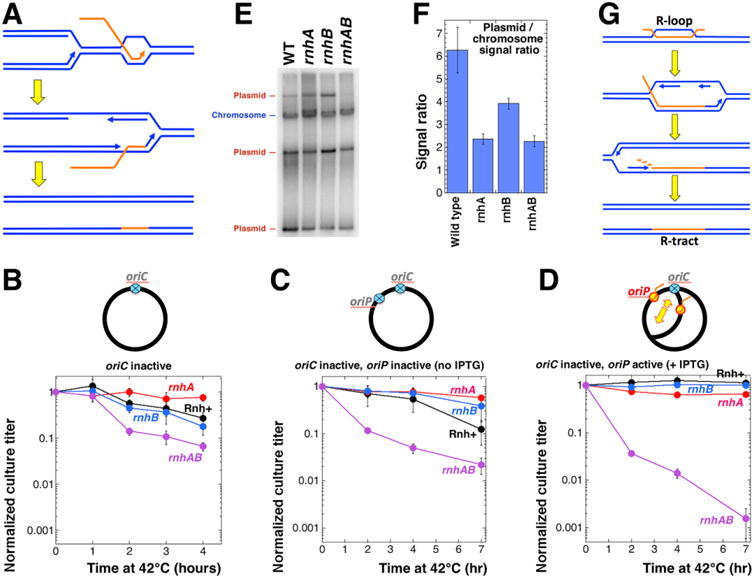
A. A model for R-tract formation due to co-directional transcript cooptation into DNA during replication in rnhAB strains. B-D All strains in these three panels are dnaA(Ts) mutants. In the chromosome schemes on the top of the corresponding graphs, inactive origins are shown as blue crossed circles, active origins are shown in red-and-yellow. The pEAK54 integration site (ColE1 origin under IPTG control) is marked as oriP. The direction of R-loop plasmid replication is identified by a bigger yellow arrow. Cultures were grown at 28°C in LB, serially diluted, spotted on LB plates (with or without IPTG) and incubated at 42°C for the indicated amount of time, then shifted to 28°C incubation for 16 hours to allow colony formation. The calculated titer of the culture for the indicated time of incubation at 42°C was normalized to the titer of the culture from the plate incubated at 28°C throughout. B. Survival of various rnh derivatives of the dnaA46(Ts) mutant after incubation at 42°C for up to 4 hours. Strains: rnh+, L-159: rnhA, L-483; rnhB, L-482; rnhAB, L-484. C. Survival of various rnh derivatives of the dnaA46(Ts) ColE1-ori mutant after incubation at 42°C for up to 7 hours without IPTG. Strains: rnh+, L-215; rnhA, L-486; rnhB, L-485: rnhAB, L-487. D. The experiment is done as in C with plates containing 1 mM IPTG for full ectopic origin induction. E. Determination of relative copy numbers of a ColE1 plasmid pAM34 in rnh strains. Total genomic DNA was isolated from the indicated strains transformed with pAM34 and grown in LB + 1 mM IPTG at 28°C to OD600 = 0.4. DNA was separated on a 1.1% agarose gel, transferred to a nylon membrane and hybridized to the pAM34 used as a probe with lacI-DNA hybridizing to the chromosome. The strains were: wild type, AB1157; rnhA, L413; rnhB, L-415; rnhAB, L-416. F. The ColE1 plasmid maintains a lower copy number per chromosome in the rnh mutants. Quantification of the relative ratio of the plasmid-to-chromosome signal from several gels like in “E”. G. A model for R-tract formation due to replication initiation from R-loop.
