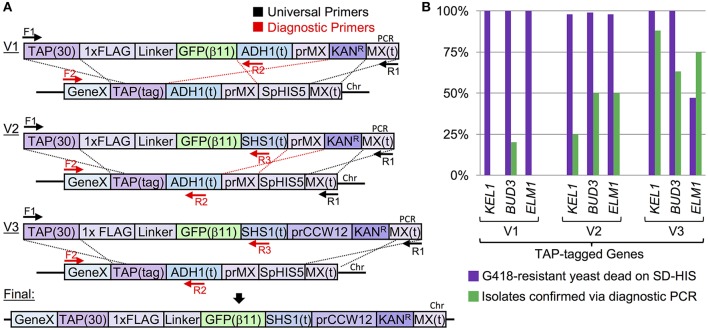
Figure 1.
Chromosomal integration of a C-terminally tagged cassette into the TAP-tagged yeast library using homologous recombination. (A) Three constructs (V1, V2, and V3) were PCR amplified (from pGF-IVL845, pGF-IVL890, and pGF-IVL985, respectively) and transformed into three yeast strains (GFY-1583, GFY-1589, and GFY-1620) containing either KEL1, BUD3, or ELM1 tagged with the TAP marker (Figure S3). These integration cassettes (Table 2) allow for a C-terminal 1xFLAG-Linker-GFP(β11) tripartite split GFP tag (Finnigan et al., 2016) to be fused to any open reading frame that is part of the TAP collection. Each PCR contains a 30 bp universal segment of the TAP linker sequence as well as the full MX terminator; black dotted lines illustrate the expected homologous sections with the chromosomal DNA. Additional identical sequences [e.g. ADH1(t)] also providing homology are illustrated with red dotted lines. Two universal primers (black arrows) amplify the common TAP linker sequence (F1, “TAP Tag clone out F”) and the MX(t) sequence (R1, “MX clone out R2”) (Table S1). Unique diagnostic primers, red arrows. Replacement of the prMX with the prCCW12 still allowed for G418 selection. (B) Quantification of the PCR integrations from (A) using both growth assays and diagnostic PCRs. G418-resistant yeast were tested on SD-HIS medium (n = 100 colonies). From SD-HIS sensitive colonies, isolates (V1, n = 10; V2/V3, n = 8) were selected and assayed by PCR as illustrated in (A). For V1, PCRs [F2, “KEL1 Internal +2908 F”/“BUD3 Internal +4381 F”/“ELM1 Internal +1455 F”; R2, “Internal ADH1(t) R”] were performed; for V2/V3, PCRs (F2/R2 and F2/R3, “SHS1(t) R”) were assayed (Table S1). At least two isolates for each integration event were confirmed via DNA sequencing.