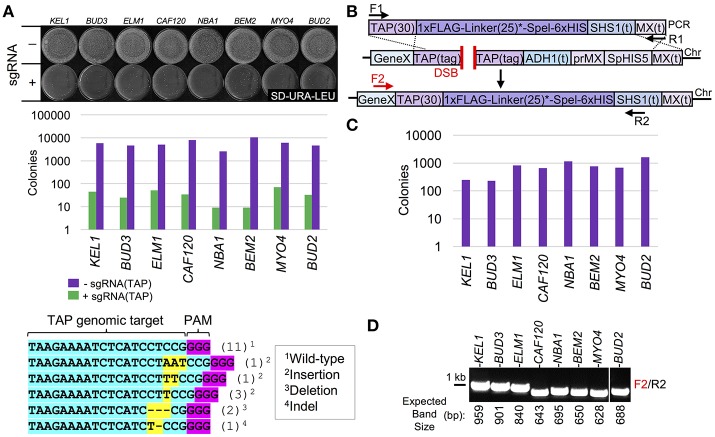
Figure 2.
Use of CRISPR/Cas9 editing to C-terminally tag the TAP haploid library. (A) Targeting of Cas9 to the TAP tag sequence at various genomic loci induces NHEJ. Eight yeast strains from the TAP collection (GFY-1583, GFY-1589, GFY-1620, GFY-2047, GFY-2056, GFY-2069, GFY-2071, and GFY-2092) were (i) transformed with a Cas9 plasmid (pGF-V789), (ii) induced in galactose for Cas9 expression, (iii) transformed with the sgRNA plasmid (pGF-V799) targeting the TAP sequence (Figure S3) or an empty pRS425 control vector, and (iv) plated to SD-URA-LEU plates (top). The total number of colonies was quantified on a log10 scale (middle). Surviving colonies from the KEL1, BUD3, and ELM1 transformation events (+sgRNA) were sequenced at their TAP-tagged loci (bottom). The number of each obtained genotype is illustrated. (B) As in Figure 1A, a C-terminal integration cassette containing a FLAG/His epitope tag and a 25-residue flexible linker (asterisk) (see Table 3) was constructed. The TAP(30) sequence contains the first 30 bp of the TAP tag cassette. (C) Strains from (A) were transformed with the Cas9 vector, the sgRNA(TAP) vector, and equimolar amounts (1,000 ng) of donor PCR DNA (F1, “TAP Tag clone out F”/R1, “MX clone out R2”), plated to SD-URA-LEU, and the total colony count quantified. (D) Colonies (n = 30–50) from (C) were selected, tested on SD-HIS medium, and a representative isolate (n = 1) was selected (lacking the S. pombe HIS5 marker) and assayed by diagnostic PCR. (F2, Gene-specific primers/R2, “SHS1(t) R”) (see Table S1). The expected PCR sizes (bp) are shown.