Abstract
Uncoupling protein 1 (UCP1) permits non-shivering thermogenesis (NST) when highly expressed in brown adipose tissue (BAT) mitochondria. Exclusive to placental mammals, BAT has commonly been regarded to be advantageous for thermoregulation in hibernators, small-bodied species, and the neonates of larger species. While numerous regulatory control motifs associated with UCP1 transcription have been proposed for murid rodents, it remains unclear whether these are conserved across the eutherian mammal phylogeny and hence essential for UCP1 expression. To address this shortcoming, we conducted a broad comparative survey of putative UCP1 transcriptional regulatory elements in 139 mammals (135 eutherians). We find no evidence for presence of a UCP1 enhancer in monotremes and marsupials, supporting the hypothesis that this control region evolved in a stem eutherian ancestor. We additionally reveal that several putative promoter elements (e.g., CRE-4, CCAAT) identified in murid rodents are not conserved among BAT-expressing eutherians, and together with the putative regulatory region (PRR) and CpG island do not appear to be crucial for UCP1 expression. The specificity and importance of the upTRE, dnTRE, URE1, CRE-2, RARE-2, NBRE, BRE-1, and BRE-2 enhancer elements first described from rats and mice are moreover uncertain as these motifs differ substantially—but generally remain highly conserved—in other BAT-expressing eutherians. Other UCP1 enhancer motifs (CRE-3, PPRE, and RARE-3) as well as the TATA box are also highly conserved in nearly all eutherian lineages with an intact UCP1. While these transcriptional regulatory motifs are generally also maintained in species where this gene is pseudogenized, the loss or degeneration of key basal promoter (e.g., TATA box) and enhancer elements in other UCP1-lacking lineages make it unlikely that the enhancer region is pleiotropic (i.e., co-regulates additional genes). Importantly, differential losses of (or mutations within) putative regulatory elements among the eutherian lineages with an intact UCP1 suggests that the transcriptional control of gene expression is not highly conserved in this mammalian clade.
Keywords: uncoupling protein 1, evolution, transcriptional regulation, enhancer, comparative analysis
Introduction
Uncoupling protein 1 (UCP1) expression is a defining characteristic of brown adipose tissue (BAT), allowing this specialized eutherian heater organ to function in non-shivering thermogenesis (NST). UCP1 spans the mitochondrial inner-membrane of brown adipocytes, acting to promote mitochondrial proton leak, which dissipates the electrochemical gradient that typically drives ATP synthase. In an effort to defend the mitochondrial proton motive force, the electron transport chain thus pumps protons into the inter-membrane space at an elevated rate via an increased level of substrate combustion, thereby resulting in substantial heat production in the form of NST (Cannon and Nedergaard, 2004; Klingenspor and Fromme, 2012).
Vital to its function, BAT is highly vascularized and localized primarily to the thoracic region, lying adjacent to major blood vessels of the heart (e.g., the Sulzer's vein) permitting effective transfer of NST heat to the rest of the body via the circulatory system (Klingenspor and Fromme, 2012; Oelkrug et al., 2015). This provides a more efficient means of heat production than shivering thermogenesis, which has major drawbacks as it impedes locomotion and produces heat in large muscle groups of the limbs that are prone to heat loss due to their high surface area to volume ratios (Oelkrug et al., 2015). For these reasons, UCP1 is widely considered to have provided a key thermoregulatory and evolutionary advantage to the eutherian lineage, particularly for small-bodied and hibernating species, and, while BAT in larger-bodied species (e.g., humans) is typically lost with the onset of adulthood, it has been generally understood to play vital role in their neonates (Cannon and Nedergaard, 2004).
The UCP1 gene predates the divergence of ray- and lobe-finned fishes (420 million years ago [MYA]) and can be distinguished from UCP2 and UCP3 paralogs by its conserved synteny among vertebrates, as UCP1 is flanked by the upstream TBC1D9 and downstream ELMOD2 loci (Jastroch et al., 2008; Klingenspor et al., 2008). UCP2 and UCP3 have been long-believed to play non-thermogenic roles, and are instead hypothesized to perform a multitude of functions including the reduction of reactive oxygen species by promoting a low level of mitochondrial proton leak when activated by fatty acids (Brand and Esteves, 2005; Echtay, 2007; Mailloux and Harper, 2011). However, a recent study by Lin et al. (2017) suggests that proton uncoupling by UCP3 permits heat production in beige adipose tissue of pigs, compensating for the loss of UCP1 in this lineage (Berg et al., 2006). Nevertheless, the functional roles of both UCP2 and UCP3 remain hotly debated. Similarly, the ancestral function of UCP1 in non-eutherians is currently unclear (Klingenspor et al., 2008). UCP1 expression has been shown to increase with cold exposure in common carp (Cyprinus carpio) brain tissue, suggesting a possible role in local thermogenesis (Jastroch et al., 2007). However, to date, this protein has not been definitively linked to heat production in ectothermic vertebrates (Jastroch et al., 2007). While the fat-tailed dunnart (Sminthopsis crassicaudata), a marsupial, displays a primitive “brownish” interscapular adipose depot that up-regulates UCP1 expression in response to cold exposure (Jastroch et al., 2008), this tissue is incapable of adaptive NST (Polymeropoulos et al., 2012) with no study demonstrating that UCP1 contributes to NST in marsupials. Although UCP1 appears to have been inactivated early in the evolution of the eutherian superorder Xenarthra (Gaudry et al., 2017), BAT-mediated adaptive thermogenesis is widely known to occur in small-bodied members of the superorders Laurasiatheria and Euarchontoglires (Oelkrug et al., 2015), and has been documented in the rock elephant shrew (Elephantulus myurus; Mzilikazi et al., 2007) and the lesser hedgehog tenrec (Echinops telfairi; Oelkrug et al., 2013), both members of the eutherian superorder Afrotheria. These observations strongly suggest that UCP1 was recruited for BAT-mediated NST in a common eutherian ancestor by gain of function mutations in the amino acid sequence of the protein and/or greater control over gene transcription that allowed highly concentrated UCP1 expression within BAT mitochondria (Klingenspor et al., 2008).
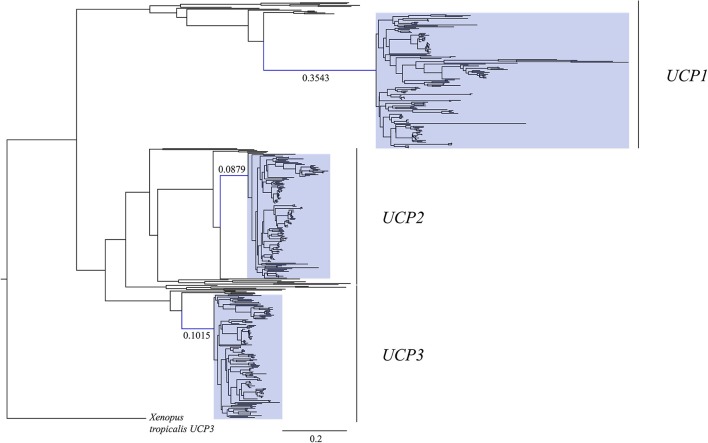
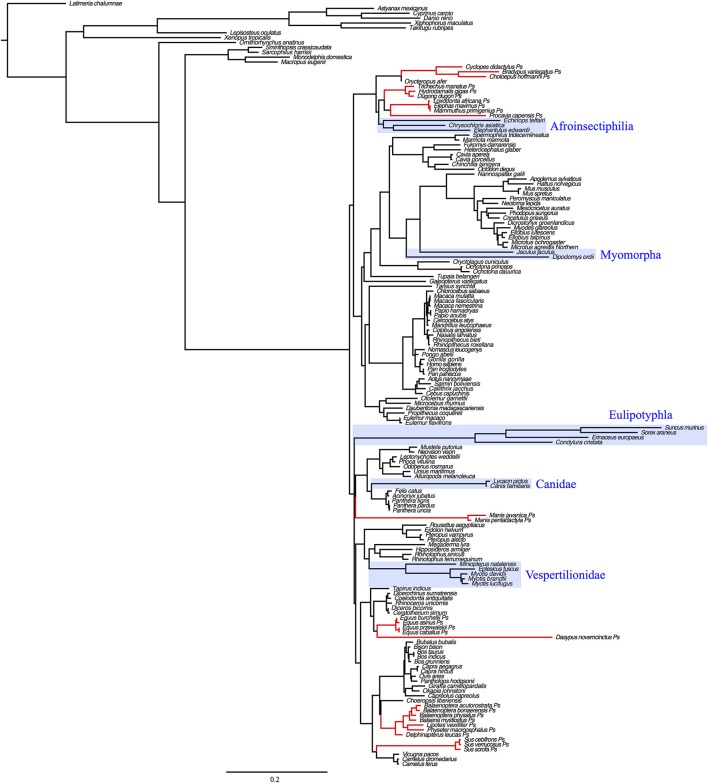
Consistent with the gain of function hypothesis, comparative phylogenetic analyses reveal that the stem eutherian branch is highly elongated in UCP1 gene trees relative to that of UCP2 and UCP3 paralogs (Saito et al., 2008; Hughes et al., 2009; Gaudry et al., 2017; Figure 1). It is thus likely that an elevated rate of non-synonymous UCP1 nucleotide substitutions in the stem eutherian branch conferred this protein with the ability to facilitate proton leak at physiologically significant levels (Jastroch et al., 2008; Klingenspor et al., 2008). While Saito et al. (2008) first proposed UCP1 evolved under positive selection in basal eutherians, more recent selection pressure analyses reveal non-synonymous to synonymous substitution ratios (dN/dS or ω) of ~0.5–0.6 that are more consistent with relaxed purifying selection (Hughes et al., 2009; Gaudry et al., 2017). However, given that UCP1 of placental mammals possess several unique amino acids relative to non-eutherians, it is possible that directional selection was limited to certain codons along the stem eutherian branch, though, so far this hypothesis remains statistically unsupported (Hughes et al., 2009; Gaudry et al., 2017).
Figure 1.
Maximum likelihood gene tree of UCP1, UCP2, and UCP3 coding sequences (N = 448) modified from Gaudry et al. (2017) to include the 16 additional species with recently available genome projects (see Table 1). The stem placental mammal branches are indicated in blue. Note that the UCP1 stem placental branch is much longer than those of UCP2 and UCP3, demonstrating a greater number of nucleotide substitutions per site. Placental mammal genes are highlighted with blue boxes. The tree was rooted with the western clawed frog (Xenopus tropicalis) UCP3.
Along with the increased rate of UCP1 evolution in stem eutherians, expression of this protein also became highly tissue-specific during the rise of BAT (Cannon and Nedergaard, 2004). In contrast to the seemingly constitutive presence of UCP1 in common carp brain, liver, and kidney tissues (Jastroch et al., 2007), eutherian UCP1 expression is tightly regulated, occurring predominantly in BAT (Cannon and Nedergaard, 2004). One notable exception, however, is the recently discovered “beige or brite (brown in white)” adipocytes in rodents (mice and rats) and humans. These are derived from white adipose cells that, upon cold exposure, become BAT-like by expressing UCP1 and by having multilocular lipid droplets and an elevated mitochondrial concentration (Harms and Seale, 2013). An important distinction in BAT (and UCP1) evolution is that BAT-dependent NST relies upon exceptionally high levels of UCP1 expression, constituting up to 10% of the mitochondrial membrane proteins, whereas UCP2 and UCP3 expression is several orders of magnitude lower (0.01–0.1%) in other tissues (Brand and Esteves, 2005). Interestingly, an enhancer box has been well documented to play a major role in eutherian UCP1 gene transcription, but is absent in the gray short-tailed opossum (Monodelphis domestica; Jastroch et al., 2008), suggesting that it originated with the advent of eutherian UCP1-mediated NST, thus highlighting the importance that gene regulation likely played in the rise of eutherian BAT-mediated thermogenesis.
Given the thermoregulatory advantages conferred by BAT, it is believed that this tissue was fundamental to the evolutionary success of eutherian mammals, and it has even been hypothesized to underlie their colonization of cold ecological niches (Cannon and Nedergaard, 2004). The documented inactivation of the UCP1 gene in suids (pigs) (Berg et al., 2006) initially emphasized the importance of BAT-mediated thermogenesis, as this inactivation appears to have had detrimental consequences as newborn piglets are widely known to have meager thermoregulatory abilities, suffering from high infant mortality when cold-stressed and relying upon shivering thermogenesis and maternal nest-building in order to maintain homeothermy (Herpin et al., 2002; Berg et al., 2006). By contrast, two recent studies (Gaudry et al., 2017; McGaugh and Schwartz, 2017) contested the conventional belief regarding the importance of BAT-mediated NST throughout the course of placental evolution. Indeed, Gaudry et al. (2017) not only detailed ancient pseudogenization events of UCP1 in eight additional eutherian lineages: Equidae (horses), Cetacea (whales and dolphins), Proboscidea (elephants and mammoths), Sirenia (sea cows), Hyracoidea (hyraxes), Pholidota (pangolins), Pilosa (sloths and anteaters), and Cingulata (armadillos), but concluded that extreme cold tolerance evolved in many of these groups in the absence of UCP1-mediated thermogenesis.
With the exception of xenarthrans and pangolins, who have adopted a strategy of reduced energy expenditure (i.e., low metabolic rates and body temperatures) associated with their low energy diets, and pigs, for which no credible explanation for UCP1 inactivation has yet been put forward, Gaudry et al. (2017) proposed that UCP1 inactivations date back to a period of substantial planetary cooling ~55 to 22 MYA that triggered pronounced increases in body size in other UCP1-lacking lineages (Gaudry et al., 2017). The inverse relationship between the surface-area-to-volume ratio and size imparts greater retention of heat in larger bodied mammals, thus larger mammals have proportionally lower rates of heat production per gram of body mass (McNab, 1983). This linkage is reflected in the diminishing fraction of eutherian body mass constituted by BAT, as well as a reduced NST capacity, with increasing body size (Heldmaier, 1971; Oelkrug et al., 2015). Heldmaier (1971) further suggested that BAT-mediated NST is negligible for mammals >10 kg. Nonetheless, several large-bodied taxa retain an intact UCP1 gene (e.g., rhinoceroses, pinnipeds, hippopotamus, and camel; Gaudry et al., 2017). Despite this finding, it remains conceivable that members of these groups do not express UCP1 in BAT, even as neonates. For example, Rowlatt et al. (1971) noted the absence of BAT upon examination of a single newborn hippopotamus (Hippopotamus amphibious), while both UCP1 expression and discernable BAT was not detected in either Weddell seal (Leptonychotes weddellii) or hooded seal neonates (Cystophora cristata) (Pearson et al., 2014). Additionally, the Bactrian camel (Camelus ferus) UCP1 gene displays a 12 base pair nucleotide deletion in exon 5 that would impart the loss of 4 amino acids in close proximity to a site that putatively binds GDP to act as a regulator (inhibitor) of protein activity (Gaudry et al., 2017). Consequently, disruptions to UCP1 regulatory regions may preclude expression of this protein in BAT of these lineages.
Evolution of eutherian UCP1 regulatory elements
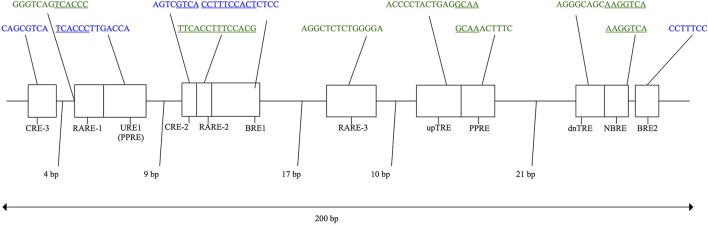
In eutherian mammals, the neuro-hormonal modulation and tissue-specific expression of UCP1 is under the control of two regulatory regions in the 5′ non-coding region of the gene—a complex distal enhancer region and a proximal promoter—through their interactions with a broad assemblage of transcription factors (Villarroya et al., 2017). Based primarily on murid rodent studies, several putative transcription factor binding motifs (see Figure 2) have been proposed within a conserved ~200 bp UCP1 enhancer box located ~2–5 kb upstream of the transcriptional start site in eutherians (Cannon and Nedergaard, 2004; Jastroch et al., 2008; Shore et al., 2012). For instance, two cAMP response elements (CREs) were discovered in mice and termed “CRE-3” and “CRE-2” (Kozak et al., 1994). CRE sites typically have a palindromic consensus sequence of 5′-T(G/T)ACGTCA-3′ (Bokar et al., 1988; Kozak et al., 1994). While the first three nucleotides of the two mouse CREs deviate from the typical consensus sequence (Figure 2), the 5′-CGTCA-3′ nucleotides remain conserved and are believed to be key for UCP1 expression. Indeed, site-directed mutagenesis of these nucleotides within the enhancer CRE of glycoprotein hormone and phosphoenolpyruvate carboxykinase genes has been shown to drastically reduce transcription factor (i.e., cAMP response element binding protein [CREB]) binding and expression in human and rat cells (Bokar et al., 1988). Two “brown adipocyte regulatory element” (BRE) protein-binding motifs (Kozak et al., 1994) also occur in the mouse UCP1 enhancer box (Figure 2). Again, site directed mutagenesis of the “TTCC” nucleotides within the BREs to a “GTAC” sequence drastically reduces UCP1 enhancer activity measured using transient expression assays (Kozak et al., 1994). In addition, Sears et al. (1996) found a stretch of nucleotides they termed “UCP regulatory element 1” (URE1), though this is referred to as the peroxisome proliferator response element (PPRE) by Jastroch et al. (2008); Jastroch also predicted a second possible PPRE motif downstream of the URE1 (PPRE) site. The URE1 motif displays high similarity to DR-1 elements (Sears et al., 1996), which are known to comprise of two direct repeats of the “AGGTCA” half-site consensus sequence separated by a single nucleotide (hence the term DR-1; i.e., direct repeats separated by 1 spacer nucleotide). In mice this sequence occurs in the reverse and complement orientation of the first DNA strand (5′- TCACCCTTGACCA-3′), and although it is not an exact match to the consensus sequence, it has been shown to bind the peroxisome proliferator-activated receptor γ and retinoid X receptor α (PPARγ-RXRα) heterodimer transcription factor (Sears et al., 1996). Conversely, mutant variants of the URE1 sequence (i.e., 5′-TCACAATTGACCA-3′ or 5′-TCACCCTAGACCA-3′) failed to bind the PPARγ-RXRα transcription factor, suggesting a key role in the functionality of the UCP1 enhancer (Sears et al., 1996). Additionally, in light of the requirement of triiodothyronine (T3) for proper BAT expression (Bianco and Silva, 1987), Rabelo et al. (1995) described two putative thyroid hormone response elements (TREs) in the rat UCP1 enhancer termed “upTRE” and “dnTRE” (Figure 2). TREs typically include two or more variations of the “AGGT(C/A)A” half-site consensus sequence separated by four nucleotides (Brent et al., 1991; Umesono et al., 1991). This same half-site sequence was mentioned above for URE1 and is indeed recognized by multiple transcription factors (Brent et al., 1991). Mutations of the 3′ portion of the upTRE (5′-AGGCAA-3′) and the dnTRE (5′-AGGTCA-3′) to “5′-ATTTAA-3′” and “5′-ATATTA-3′”, respectively, eliminate T3 receptor interactions with the rat UCP1 enhancer (Rabelo et al., 1995). Three putative retinoic acid response elements (RAREs) within the rat UCP1 enhancer have also been described by Rabelo et al. (1996), though both RARE-1 and RARE-2 overlap with other binding motifs (see Figure 2). Nonetheless, mutations increasing the AT-richness of these former regulatory elements were shown to significantly disrupt retinoic acid receptor (RAR) and retinoid X receptor (RXR) transcription factor binding (Rabelo et al., 1996). Finally, Kumar et al. (2008) noted a putative nerve growth factor response element (NBRE) within the UCP1 enhancer of mice (Figure 2) that binds nuclear receptors 4A (NR4A), which acts to promote gene transcription. In addition to the enhancer box, Shore et al. (2012) described a 678 bp putative regulatory region (PRR) located 2,095 bp upstream of the transcriptional start site in humans that was conserved in 14 of 25 of the eutherian species they examined. While Shore et al. (2012) found no evidence that this conserved region plays a role in UCP1 expression, they did note that it encompassed several possible transcription factor binding motifs, including DR1, DR3, DR4, CEBP (CCAAT-enhancer-binding proteins), CREB, and PPAR.
Figure 2.
Schematic of the murid UCP1 enhancer with putative transcription factor binding motifs shown for the rat (green) and mouse (blue) based on a combination of previous studies (see text for details). Regions of overlap between adjacent transcription factor motifs are underlined.
Transcriptional control of the UCP1 gene has also been hypothesized to be regulated by a basal promoter occurring within ~250 bp upstream of the transcription start site (Shore et al., 2010). Within this region, Bouillaud et al. (1988) identified a putative TATA box and a CCAAT binding site located ~20 and ~30 bp upstream of the transcriptional start site of the rat UCP1 gene, respectively. Generally, the TATA box consists of an A/T-rich consensus sequence (5′-TATAAAA-3′; Xu et al., 1991) that interacts with the TATA binding protein (TBP), one of the components of the transcription factor IID (TFIID) that initiates transcription via RNA polymerase II (Nakajima et al., 1988; Patikoglou et al., 1999). The promoters of some mammalian genes (e.g., globins) also contain a CCAAT box typically situated –60 to –100 bp upstream of the transcription start site that binds nuclear transcription factor Y (NF-Y) subunit or CCAAT/enhancer binding protein (C/EBP), which then aids in the initiation of transcription via RNA polymerase II (Mantovani, 1999). Additionally, a putative CRE site (termed CRE-4) occurs ~130 bp upstream of the mouse UCP1 transcriptional start site in a reverse and compliment orientation (5′-TGACGCGC-3′), with mutations to this sequence eliminating 90–95% of reporter gene expression (Kozak et al., 1994). Yubero et al. (1994) further noted three GCCCCT sequences occurring within ~210 bp of the transcriptional start site of the rat, which DNAse 1 footprinting analyses suggest interact with nuclear proteins found within BAT cells, but these have not been defined as protein binding motifs.
Finally, a CpG island surrounding the UCP1 proximal promoter and extending into exon 1 has been described in several eutherian species (Kiskinis et al., 2007; Shore et al., 2010, 2012). CpG islands contain high densities of cytosine (C) and guanine (G) nucleotide pairs occurring in the 5′ to 3′ direction and linked by a phosphate (i.e., 5′–C–phosphate–G–3′). These CpG dinucleotides are uncommon in vertebrate genomes, typically occurring at only 20–25% of the frequency anticipated by random chance and act as DNA methylation sites that can modulate gene transcription (Gardiner-Garden and Frommer, 1987). Located immediately upstream of many housekeeping genes, CpG islands are believed to play a major role in their transcriptional control (Gardiner-Garden and Frommer, 1987). Indeed, methylation of CpG dinucleotides immediately upstream of the UCP1 gene have been shown to modulate gene activity by blocking transcription, whereas demethylation promotes transcription (Shore et al., 2010). Thus, this CpG island has been postulated to be important for UCP1 gene regulation and, potentially, tissue specific expression within BAT (Kiskinis et al., 2007; Shore et al., 2010).
Because the majority of studies investigating the transcriptional control of UCP1 have focused on rodents, the status of these transcription factor binding motifs in other eutherian species remain largely unexplored. Here we use genome mining and hybridization-capture techniques coupled with next-generation sequencing to identify and examine UCP1 transcriptional regulatory elements in 139 mammals (135 eutherians). Briefly, putative transcription factor binding motifs and CpG islands were evaluated using a comparative approach to first determine if they are universally conserved among eutherian superorders with functional BAT, and second to test if they are mutated or lost in large-bodied species that presumably have little or no need for NST. We further anticipated that crucial DNA motifs involved in UCP1 transcription would have deteriorated via millions of years of neutral evolution in the nine lineages for which UCP1 has been inactivated.
Materials and methods
UCP1 regulatory sequences
In total, UCP1 upstream regions of 139 mammals (1 monotreme, 3 marsupials, 3 xenarthrans, 11 afrotherians, 65 laursiatherians, and 56 euarchontoglires) were examined for transcriptional regulatory elements (see Table 1 for species list). This data set employed 116 species whose UCP1 loci were previously annotated by Gaudry et al. (2017) together with 16 additional species whose genomes have recently been sequenced (denoted by asterisks in Table 1). Regulatory elements of seven additional eutherians were also retrieved by hybridization capture and next-generation sequencing techniques. Briefly, UCP1 enhancers, PRRs, and basal promoters of four rhinoceroses (black rhinoceros: Diceros bicornis, Indian rhinoceros: Rhinoceros unicornis, Sumatran rhinoceros; Dicerorhinus sumatrensis, and woolly rhinoceros; Coelodonta antiquitatis), one tapir (Malayan tapir; Tapirus indicus), and two sirenians (dugong; Dugong dugon, and Steller's sea cow; Hydrodamalis gigas), were targeted using hybridization capture and next-generation sequencing techniques (Springer et al., 2015; Gaudry et al., 2017). Barcoded rhinoceros DNA libraries were constructed using NEBNext Fast DNA Library Prep Set for Ion Torrent and NEBNext DNA Library Prep Master Mix Set for 454 kits (New England Biolabs; Ipswich, Massachusetts, USA) and target-enriched using MyBaits (Mycroarray; Ann Arbor, Michigan, USA) 120mer RNA probes designed to capture UCP1 exons and regulatory elements based on the orthologous sequences of the white rhinoceros (Ceratotherium simum) genome. The captured rhinoceros reads were sequenced on an Ion Torrent PGM platform using Ion 314 v2 and Ion 318 v2 barcoded chips and an Ion PGM Hi-Q sequencing kit (Applied Biosystems; Foster City, California, USA). Sirenian DNA libraries prepared following the methods of Meyer and Kircher (2010) were enriched using an Agilent SureSelect Capture array with probes designed from African elephant (Loxodonta africana) UCP1 upstream sequences. Sirenian DNA reads were sequenced on Illumina GAIIx and HiSeq2500 (Illumina Inc.; San Diego, California, USA) platforms. Sequenced reads were assembled to reference sequences of the white rhinoceros or manatee (Trichechus manatus) using the “map to reference” feature in Geneious R9.1 (Biomatters Ltd.; Auckland, New Zealand) at 20% maximum mismatch per read and consensus sequences were generated.
Table 1.
Presence and absence of the UCP1 enhancer, putative regulatory region (PPR), and CpG island in 139 mammalian species.
| Species name | Enhancer | PPR | CPG island | Accession number |
|---|---|---|---|---|
| MONTREMATA | ||||
| Ornithorhynchus anatinus | X | X | X | NW_001794248.1 |
| MARSUPIALIA | ||||
| Monodelphis domestica | X | X | X | AAFR03015618.1 |
| Macropus eugenii | / | / | X | ABQO020217652.1 |
| Sarcophilus harrisii | / | / | X | AEFK01228715.1 |
| XENARTHRA | ||||
| Choloepus hoffmanni | X | / | / | |
| Dasypus novemcinctus | X | X | Yes | AAGV03181320.1 |
| Mylodon darwinii | Yes | / | / | SRX327588 |
| AFROTHERIA | ||||
| Chrysochloris asiatica | Yes | Yes | X | AMDV01244955.1 |
| Dugong dugon | Yes | Yes | / | MF871621 |
| Echinops telfairi | Yes | Yes | Yes | AAIY02209271.1 |
| Elephantulus edwardii | Yes | Yes | X | AMGZ01097263.1 |
| Elephas maximus | Yes | Yes | Yes | SRX1015608; SRX1015606; SRX1015604; SRX1015603 |
| Hydrodamalis gigas | Yes | / | / | MF871622 |
| Loxodonta africana | Yes | Yes | Yes | AAGU03034821.1 |
| Mammuthus primigenius | Yes | Yes | / | SRX1015727; SRX1015732; SRX1015743; SRX1015748; SRX001906; ERP008929 |
| Orycteropus afer | Yes | Yes | X | ALYB01104541.1 |
| Procavia capensis | X | Yes | Yes | ABRQ02143236.1 |
| Trichechus manatus latirostris | Yes | Yes | Yes | AHIN01109623.1 |
| LAURASIATHERIA | ||||
| Acinonyx jubatus | Yes | Yes | Yes | LLWD01000416.1 |
| Ailuropoda melanoleuca | Yes | Yes | Yes | LNAT01000144.1 |
| Balaena mysticetus | Yes | Yes | Yes | SRX790318, SRX790317, SRX790316, SRX790303, SRX790319 |
| Balaenoptera acutorostrata | Yes | Yes | Yes | ATDI01065547.1 |
| Balaenoptera bonaerensis | Yes | Yes | Yes | BAUQ01197845.1 |
| Balaenoptera physalus | Yes | Yes | Yes | SRX1571086, SRX323050 |
| Bison bison | Yes | Yes | Yes | JPYT01100523.1 |
| Bos grunniens | Yes | Yes | Yes | AGSK01075302.1 |
| Bos indicus | / | Yes | / | AGFL01142554.1 |
| Bos taurus | Yes | Yes | Yes | DAAA02044420.1 |
| Bubalus bubalis | Yes | Yes | Yes | AWWX01630119.1 |
| Camelus dromedarius * | Yes | Yes | Yes | LSZX01012659.1 |
| Camelus ferus | Yes | Yes | Yes | AGVR01051296.1; AGVR01051297.1 |
| Canis lupus familiaris | Yes | X | / | AAEX03011713.1 |
| Capra aegagrus | Yes | Yes | Yes | CBYH010071014.1 |
| Capra hircus | Yes | Yes | / | AJPT01162992.1; AJPT01162993.1 |
| Capreolus capreolus | Yes | Yes | Yes | CCMK010092645.1; CCMK010104759.1 |
| Ceratotherium simum | Yes | Yes | Yes | AKZM01017598.1 |
| Coelodonta antiquitatis | Yes | Yes | Yes | MF871623 |
| Condylura cristata | Yes | X | X | AJFV01047153.1 |
| Dicerorhinus sumatrensis | Yes | Yes | Yes | MF871625 |
| Diceros bicornis | Yes | Yes | Yes | MF871624 |
| Eidolon helvum | Yes | Yes | / | AWHC01286101.1; AWHC01029981.1 |
| Eptesicus fuscus | Yes | X | Yes | ALEH01005956.1 |
| Equus asinus | Yes | Yes | Yes | JREZ01000001.1 |
| Equus caballus | Yes | Yes | / | AAWR02018850.1; AAWR02018851.1 |
| Equus przewalskii | Yes | Yes | Yes | ATBW01036321.1; ATBW01036322.1 |
| Erinaceus europaeus | / | / | X | AMDU01193160.1; AMDU01193161.1; AMDU01193162.1 |
| Felis catus | Yes | Yes | Yes | AANG02062919.1 |
| Giraffa camelopardalis * | Yes | Yes | X | LVKQ01071482.1 |
| Hipposideros armiger * | Yes | Yes | Yes | NW_017731683.1 |
| Leptonychotes weddellii | Yes | Yes | Yes | APMU01115165.1; APMU01141180.1 |
| Lipotes vexillifer | Yes | Yes | Yes | AUPI01000024.1 |
| Lycaon pictus * | Yes | X | / | LPRB01000019.1 |
| Manis javanica * | X | X | X | NW_016530114.1 |
| Manis pentadactyla | X | X | X | JPTV01131901.1 |
| Megaderma lyra | Yes | / | / | AWHB01167753.1; AWHB01348443.1; AWHB01348444.1 |
| Miniopterus natalensis * | Yes | X | Yes | NW_015504404.1 |
| Mustela putorius furo | Yes | Yes | Yes | AGTQ01041845.1 |
| Myotis brandtii | Yes | X | Yes | ANKR01273867.1; ANKR01273868.1 |
| Myotis davidii | Yes | X | Yes | ALWT01125743.1 |
| Myotis lucifugus | Yes | X | Yes | AAPE02001462.1 |
| Odobenus rosmarus | Yes | Yes | Yes | ANOP01028105.1 |
| Okapia johnstoni * | Yes | Yes | Yes | LVCL010093660.1; LVCL010093662.1 |
| Orcinus orca | X | X | X | ANOL02004931.1 |
| Ovis aries | Yes | Yes | Yes | AMGL01037664.1; JN604985.1 |
| Panthera pardus * | Yes | Yes | Yes | NW_017619848.1 |
| Panthera tigris altaica | Yes | Yes | Yes | ATCQ01112915.1 |
| Panthera uncia | Yes | Yes | / | SRX273036 |
| Pantholops hodgsonii | Yes | Yes | Yes | AGTT01188813.1 |
| Physeter macrocephalus | X | Yes | Yes | AWZP01062081.1 |
| Pteropus alecto | Yes | Yes | Yes | ALWS01011689.1 |
| Pteropus vampyrus | Yes | Yes | Yes | ABRP02126915.1 |
| Rhinoceros unicornis | Yes | Yes | Yes | MF871626 |
| Rhinolophus ferrumequinum | Yes | Yes | Yes | AWHA01040305.1 |
| Rhinolophus sinicus * | Yes | Yes | Yes | NW_017738992.1 |
| Rousettus aegyptiacus * | Yes | Yes | Yes | NW_015494583.1 |
| Sorex araneus | Yes | X | Yes | AALT02056093.1 |
| Sus cebrifrons | Yes | / | / | ERX953604-ERX953626; ERX149172 |
| Sus scrofa | Yes | X | X | LUXQ01106311.1 |
| Sus verrucosus | Yes | / | / | ERX1054048-ERX1054067; ERX149174 |
| Tapirus indicus | Yes | / | Yes | MF871627 |
| Tursiops truncatus | X | X | X | ABRN02199412.1 |
| Ursus maritimus | Yes | Yes | / | AVOR01014285.1; AVOR01014286.1 |
| Vicugna pacos | Yes | Yes | Yes | ABRR02134987.1; ABRR02134989.1 |
| EUARCHONTOGLIRES | ||||
| Aotus nancymaae | Yes | Yes | Yes | JYKP01215429.1 |
| Apodemus sylvaticus | Yes | / | X | LIPJ01452544.1; LIPJ01184746.1; LIPJ01447868.1; LIPJ01014497.1 |
| Callithrix jacchus | Yes | Yes | Yes | ACFV01002817.1 |
| Cavia aperea | / | / | / | AVPZ01000778.1 |
| Cavia porcellus | Yes | Yes | Yes | AAKN02011801.1 |
| Cebus capuchinis * | Yes | Yes | Yes | NW_016107319.1 |
| Cercocebus atys | Yes | Yes | Yes | JZLG01060688.1 |
| Chinchilla lanigera | Yes | Yes | Yes | AGCD01027651.1 |
| Chlorocebus sabaeus | Yes | Yes | Yes | AQIB01017419.1 |
| Colobus angolensis | Yes | Yes | Yes | JYKR01122839.1 |
| Cricetulus griseus | Yes | Yes | X | AFTD01128393.1; AFTD01128394.1 |
| Daubentonia madagascariensis | Yes | Yes | / | AGTM011584638.1; AGTM011584996.1; AGTM011708528.1; AGTM012010142.1; AGTM011594144.1 |
| Dipodomys ordii | Yes | X | Yes | ABRO02057411.1 |
| Ellobius lutescens * | Yes | X | Yes | LOEQ01000193.1 |
| Ellobius talpinus * | Yes | X | Yes | LOJH01032235.1 |
| Eulemur flavifrons | Yes | Yes | Yes | LGHW01000184.1 |
| Eulemur macaco | Yes | Yes | Yes | LGHX01000184.1 |
| Fukomys damarensis | Yes | Yes | Yes | AYUG01151056.1 |
| Galeopterus variegatus | Yes | Yes | / | JMZW01045215.1; JMZW01045216.1 |
| Gorilla gorilla gorilla | Yes | Yes | Yes | NW_004002547.1 |
| Heterocephalus glaber | Yes | Yes | Yes | AFSB01162372.1; AFSB01162373.1 |
| Homo sapiens | Yes | Yes | Yes | NG_012139.1 |
| Jaculus jaculus | Yes | Yes | Yes | AKZC01091543.1 |
| Macaca fascicularis | Yes | Yes | Yes | CAEC01514737.1 |
| Macaca mulatta | Yes | Yes | Yes | AANU01271750.1 |
| Macaca nemestrina | Yes | Yes | Yes | JZLF01028562.1 |
| Mandrillus leucophaeus | Yes | Yes | Yes | JYKQ01107154.1; JYKQ01107155.1 |
| Marmota marmota | Yes | Yes | Yes | CZRN01000015.1 |
| Mesocricetus auratus | Yes | X | Yes | APMT01116524.1; NM_001281332.1 |
| Microcebus murinus | Yes | Yes | Yes | ABDC01082367.1 |
| Microtus agrestis | Yes | X | Yes | LIQJ01004042.1 |
| Microtus ochrogaster | Yes | X | X | AHZW01157105.1; AHZW01157106.1 |
| Mus musculus | Yes | X | X | CAAA01024310.1 |
| Mus spretus * | Yes | X | X | LVXV01001867.1 |
| Myodes glareolus | Yes | / | / | LIPI01003929.1 |
| Nannospalax galili | Yes | Yes | X | AXCS01128925.1 |
| Nasalis larvatus | Yes | Yes | Yes | JMHX01319533.1 |
| Neotoma lepida * | / | / | X | LZPO01075894.1 |
| Nomascus leucogenys | Yes | Yes | Yes | ADFV01177960.1 |
| Ochotona princeps | Yes | X | X | ALIT01060999.1 |
| Octodon degus | Yes | Yes | Yes | AJSA01193669.1; AJSA01193670.1; AJSA01193671.1 |
| Oryctolagus cuniculus | Yes | Yes | Yes | AAGW02045633.1 |
| Otolemur garnettii | Yes | Yes | X | AAQR03074138.1 |
| Pan paniscus | Yes | Yes | Yes | AJFE01070904.1 |
| Pan troglodytes | Yes | Yes | / | AACZ03032212.1; AACZ03032213.1 |
| Papio anubis | Yes | Yes | Yes | AHZZ01043343.1 |
| Peromyscus maniculatus | Yes | X | X | AYHN01134223.1 |
| Pongo abelii | Yes | Yes | Yes | ABGA01062109.1 |
| Propithecus coquereli | Yes | Yes | Yes | JZKE01017273.1 |
| Rattus norvegicus | Yes | X | X | AAHX01097782.1 |
| Rhinopithecus bieti * | Yes | Yes | Yes | NW_016805762.1 |
| Rhinopithecus roxellana | Yes | Yes | Yes | JABR01098768.1 |
| Saimiri boliviensis | Yes | Yes | Yes | AGCE01051213.1 |
| Spermophilus tridecemlineatus | Yes | Yes | Yes | AGTP01049378.1 |
| Tarsius syrichta | Yes | Yes | / | ABRT02355486.1 |
| Tupaia belangeri chinensis | Yes | Yes | Yes | ALAR01031045.1 |
Xs = absent, / = inconclusive due to insufficient data,
= 16 species with recently published genome projects since the Gaudry et al. (2017) publication. Accession numbers are also provided for contigs and SRA projects.
For publically available genomes, UCP1 regulatory sequences were acquired using genome-mining techniques of sequences available on the National Center for Biotechnology Information web server. UCP1-containing contigs were first acquired by performing nucleotide BLAST searches employing the “discontinuous megablast” option against whole genome shotgun (WGS) contigs of mammalian genome projects using human UCP1 CDS (NM_021833.4) as a query. If the contigs did not extend ~5 kb upstream of the UCP1 transcriptional start site to include the enhancer box, an additional nucleotide BLAST was performed using the human UCP1 enhancer sequence as a query. For several species with genome projects that have not yet been fully assembled (e.g., Sus cebifrons, Sus verrucosus, Elephas maximus, Mammuthus primigenius, Balaena mysticetus, Balaenoptera physalus, Mylodon darwinii, Panthera unica), short read archive (SRA) BLASTs were performed in order to obtain the UCP1 regulatory elements. Contigs from top BLAST hits were then imported into Sequencher v5.1 (Gene Codes Corporation; Ann Arbor, Michigan, USA) and the exons and regulatory regions annotated by aligning orthologous human UCP1 sequences (exons 1–6 and enhancer), initially at a 85% minimum match percentage. If the sequences were too divergent to assemble at that stringency, the minimum match percentage was progressively decreased to 60% or until the sequences successfully assembled. UCP1 coding regions for the 16 species not included in the Gaudry et al. (2017) study were also examined for the presence of inactivating (e.g., splice site, frameshift, and non-sense) mutations.
The PRR proposed by Shore et al. (2012) was generally less conserved than the enhancer, often with large insertions or deletions, therefore the same annotation methods described above could not be effectively applied to this region. Instead, dot plots were performed in Geneious R9.1 (Biomatters Ltd.) which uses the EMBOSS 6.5.7 dotmatcher tool to compare sequence identities of the human PRR vs. the upstream sequence of other mammalian species using a window size of 25, a threshold of 45, and the high sensitivity setting with a probabilistic scoring matrix. The PRR was determined to be present if a conserved region >100 bp relative to the human sequence was discernible from the dot plots. The boundaries of the PRRs were estimated using the dot plot and annotated. The PRRs of species listed in Table 2 were then screened in rVista 2.0 (Loots and Ovcharenko, 2004) for the presence of putative transcription factor binding motifs [DR1, DR3, DR4, CEBP (CCAAT-enhancer-binding proteins), CREB, and PPAR] shared with humans, as performed by Shore et al. (2012). Insertions larger than 100 bp relative to the human PRR were removed prior to screening in rVista using the vertebrate TRANSFAC professional V10.2 library with the “matrix similarity optimized for function” setting.
Table 2.
Possible transcription factor binding motifs within the PRR of selected species screened using rVista 2.0.
| Species | Motif | Position | Sequence |
|---|---|---|---|
| Homo sapiens | CREB | 24+ | catggCATCAgttc |
| DR3 | 227− | cagaGGTTCACTAGAGTCaac | |
| DR4 | 230− | agGTTCACTAGAGTCAa | |
| Marmota marmota | PPAR_DR1 | 50− | tGGTCAAAGGACt |
| DR4 | 326− | tgGGTCCCTTAAGGTCa | |
| DR1 | 393− | TGACACTTATCCc | |
| Oryctolagus cuniculus | CREB | 373− | ccTAACATCAcc |
| CEBP | 519− | gcTCCATTGCCTAACTCt | |
| PPAR_DR1 | 592+ | tGGCCCTTGGCCc | |
| PPAR_DR1 | 601+ | gCCCCTTTGTCCc | |
| Camelus ferus | CEBP | 271− | taTACATTTGGGCATACt |
| CEBP | 503− | tgTTCCTTTCCTAATTGt | |
| CREB | 636− | tgtCATCAcct | |
| Bos taurus | CREB | 149+ | CGTCAg |
| CEBP | 240− | taTGCATTATAACAAACa | |
| CEBP | 471− | tgTTTCTTTCCTAATTTg | |
| PPAR_DR1 | 487+ | tGACCTTTGATAa | |
| PPAR_DR1 | 542+ | tGACCCTTGACCc | |
| Giraffa camelopardalis | CREB | 150+ | CGTCAg |
| CREB | 476− | tgTTTCTTTCCTAATTTg | |
| PPAR_DR1 | 492+ | tGACCTTTGATAa | |
| PPAR_DR1 | 547+ | tGACCCTTGACCc | |
| Balaenoptera acutorostrata | DR1 | 96+ | aGGGGAAGGGACA |
| CEBP | 518− | taTTTCTTTCCTAACTTt | |
| PPAR_DR1 | 587+ | tGGCCCTTGACCc | |
| DR1 | 587− | TGGCCCTTGACCc | |
| DR1 | 594− | TGACCCCTTTCCc | |
| Lipotes vexillifer | DR3 | 291+ | accGAACATTCTCAATCtgct |
| CEBP | 509− | taTTTCTTTCCTAACTTt | |
| PPAR_DR1 | 580+ | tGGCCCTTGACCc | |
| DR1 | 587− | TGACCCCTTTCCc | |
| Ceratotherium simum | DR1 | 108+ | aGGGGAAGGGACA |
| DR4 | 246− | agGATCACTAGAGTTAg | |
| CEBP | 284− | taTACATTTAGTCATACt | |
| DR3 | 304+ | accGAACATTCTCAATCtctg | |
| DR4 | 425+ | tGTCCTCTTTTGACAtt | |
| PPAR_DR1 | 453+ | tCACACTTGACCc | |
| Equus przewalskii | CEBP | 9+ | cTTTCACAAtcc |
| CREB | 36− | caTAGCGTCAgt | |
| CREB | 41+ | CGTCAg | |
| DR4 | 234− | agGTTCACTAGAGTTAg | |
| PPAR_DR1 | 537+ | tTACCTTTGACCa | |
| DR1 | 592− | TGGTCCTTGACCc | |
| CREB | 667+ | ttGCTGACTccc | |
| Equus caballus | DR4 | 224− | agGTTCACTAGAGTTAg |
| PPAR_DR1 | 524+ | tTACCTTTGACCa | |
| DR1 | 579− | TGGTCCTTGACCc | |
| CREB | 654+ | ttGCTGACTccc | |
| Pteroupus vampyrus | CREB | 37+ | catagCATCAgctc |
| DR4 | 408+ | tGTCCTCTTTTGACAtt | |
| PPAR_DR1 | 575+ | tGGCCCTTGACCc | |
| DR1 | 582− | TGACCCCTTTCCt | |
| Ailuropoda melanoleuca | DR1 | 85+ | aGGGGAAGGGACA |
| CREB | 505+ | ttGATGAGGccc | |
| DR1 | 554− | TGGCCCATGACCc | |
| PPAR_DR1 | 561+ | tGACCCTTTGCCt | |
| CREB | 628+ | ttGCTGACTccc | |
| Odobenus rosmarus | DR1 | 92+ | aGGGGAAGGGACA |
| DR4 | 406+ | tGTCCTCTTTTGACAtt | |
| DR1 | 567− | TGGCCCATGACCc | |
| PPAR_DR1 | 574+ | tGACCCTTTTCCt | |
| CREB | 670+ | ttGCTGACTccc | |
| Panthera pardus | DR4 | 240+ | tGTCCTCTTTTGACAca |
| Leptonychotes weddellii | DR1 | 90+ | aGGGGAAGGGACA |
| DR4 | 403+ | tGTCCTCTTTTGACAtt | |
| DR1 | 564− | TGGCCCATGACCc | |
| PPAR_DR1 | 571+ | tGACCCTTTTCCt | |
| CREB | 671+ | ttGCTGACTccc | |
| Procavia capensis | CREB | 59− | ccTAACATCAcc |
| DR1 | 273− | TGGTCCTTGACCt | |
| CREB | 278+ | cttgaCCTCAttgc | |
| CREB | 280+ | TGACCTca | |
| Loxodonta africana | CREB | 32+ | acataCATCAgctc |
| CREB | 347− | caTAACATCAcc | |
| CREB | 424− | tTGACG | |
| PPAR_DR1 | 566+ | tGGCCCTTGACCc | |
| Trichechus manatus | CREB | 140− | tgAGGTCA |
| CREB | 369− | taaCATCACCaa | |
| PPAR_DR1 | 587+ | tGGCCCTTGACCc | |
| Echinops telfairi | PPAR_DR1 | 189− | gGGTCAAGGATCa |
| CREB | 326− | ccTGACATCAct |
Duplicates sites were removed. Position is indicated relative to the start of the PRR sequence and the strand is indicated with + or − symbols.
Basal promoter regions were identified by performing alignments of 600 bp upstream of the ATG start codon for each species with available sequence data. The rat and mouse upstream sequences contain several putative promoter motifs (e.g., TATA box, CCAAT site, CRE-4, and GCCCCT sites) and thus were used as reference sequences. CpG islands within the 5′ region of UCP1 were identified using the EMBOSS CpGplot tool (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/). Kiskinis et al. (2007) noted that the UCP1 CpG island occurs immediately upstream of the UCP1 open reading frame but may also extend into exon 1, therefore, 1 kb upstream of exon 2 was screened for the presence of CpG islands. EMBOSS CpGplot positively identifies CpG islands if a sequence >200 bp contains an observed/expected ratio of CpGs exceeding 0.6, with a GC content >50%, meeting the criteria proposed by Gardiner-Garden and Frommer (1987). The default window size of 100 bp was used for these runs.
The UCP1 genes of non-eutherian mammals were also examined for the presence or absence of regulatory elements. Contigs of the Tasmanian devil (Sarcophilus harrisii) and Tammar wallaby (Macropus eugenii) were too short to encompass a potential enhancer occurring ~5 kb upstream of the transcriptional start site. However, contigs of the platypus (Ornithorhynchus anatinus) and gray short-tailed opossum were sufficiently long to create dot plots of the upstream sequence in order to screen for homologous regulatory elements occurring in the human. Some eutherian species displayed inactivated UCP1 genes with deletions of whole exons (e.g., Chinese pangolin; Manis pentacatyla, Javan pangolin; Manis javanica, nine-banded armadillo; Dasypus novemcinctus), or deletion of the entire gene (killer whale and bottlenose dolphin). The annotation techniques described above did not reveal the presence of a UCP1 enhancer in these species; thus, sequence identity comparisons against human UCP1 were performed using Easyfig 2.1 (Sullivan et al., 2011). This analysis was also performed for the rat and cow (Bos taurus) since these were species are known to display UCP1 enhancers while the cow also contains a PRR region (Shore et al., 2012).
Finally, regions containing enhancer and basal promoter sequences for each species were imported into Geneious 9.1 and multispecies nucleotide alignments were generated using the MUSCLE alignment tool (Edgar, 2004) with default settings. A consensus eutherian sequence representing the simple majority (>50%) was generated from this dataset based only on species for which the UCP1 gene is intact (i.e., species with documented UCP1 pseudogenes (Gaudry et al., 2017) were not included in the consensus calculations). For some eutherian species, pairwise alignments were also created against the human enhancer to obtain the percent sequence identity values. Conserved motifs and putative transcription factor binding sites were annotated. Recognized transcription factor binding motifs within the UCP1 enhancer (illustrated in Figure 2) were examined by eye in each eutherian species and scrutinized for mutations that potentially affect DNA-protein interactions based on previous site directed mutagenesis studies. Additionally, the consensus enhancer region sequence (see above), together of those of seven species spanning the three mammalian superorders for which UCP1 is intact, were screened for the presence of all vertebrate transcription factors in the TRANSFAC professional V10.2 library using rVista with the “matrix similarity optimized for function” setting.
Phylogenetic trees
To generate a combined UCP1, UCP2, and UCP3 coding sequence phylogenetic tree, the data set of Gaudry et al. (2017) was updated to include coding sequences of the 16 additional species with recently published genomes (Table 1). The resulting 448 UCP genes were aligned using MUSCLE (Edgar, 2004), and a maximum likelihood tree constructed using RAxML (Randomized Axelerated Maximum likelihood) version 7.2.8 (Stamatakis, 2006) with the “GTR Gamma” nucleotide model and “rapid bootstrapping and search for best scoring tree” setting. The program was performed for 500 bootstrap replicates.
In order to trace the evolutionary gain and loss of UCP1 transcriptional regulatory elements, we also constructed a 41-gene species tree for the 139 mammals included in this study following the methods of Gaudry et al. (2017). Briefly, this data set included coding and non-coding sequences from 30 nuclear (A2AB, ADRB2, APP, ATP7A, ADORA3, APOB, BCHE, BDNF, BMI1, BRCA1, BRCA2, CHRNA1, CMYC, CNR1, CREM, DMP1, ENAM, EDG1, FBN1, GHR, IRBP, MC1R, PLCB4, PNOC, RAG1, RAG2, SWS1, TTN, TYR1, VWF) and 11 mitochondrial loci (12S rRNA, 16S rRNA, CYTB, COI, COII, COIII, ND1, ND2, ND3, ND4, ND5). A 50,911 bp concatenated supermatrix was aligned in MUSCLE. The supermatrix was divided into 32 partitions (see Supplementary Materials). Each nuclear gene was assigned an individual partition, while 12S rRNA and 16S rRNA were combined to create one partition, and the nine remaining mitochondrial genes were also combined into a single partition. An independent GTR Gamma model was estimated for of these partitions and a maximum likelihood tree was generated in RAxML 7.2.8 using the same settings described above with 100 bootstrap replicates.
Results
UCP1 coding sequences
All of the 16 newly acquired UCP1 CDSs were intact with the exception of the Javan pangolin, which displays the same mutations as the Chinese pangolin pseudogene (i.e., frameshift, splice site and non-sense mutations, deletion of exons 1 and 2) documented by Gaudry et al. (2017). Similarly, the 12 bp deletion that calls into question the functionality of the Bactrian camel UCP1 gene (Gaudry et al., 2017) is also present in the dromedary camel (Camelus dromedarius). Conversely, the UCP1 CDS of the giraffe (Giraffa camelopardalis) is intact, despite its large body size.
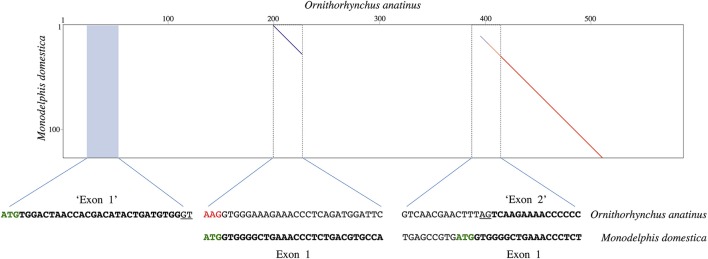
The predicted platypus UCP1 CDS available on GenBank (accession number: XM_001512650) is unique in that it creates a hypothetical open reading frame composed of seven exons; the usual 126 bp exon 1 is divided into two separate exons of 30 and 120 bp in length. The placement of these putative exons are displayed in a dot plot comparison with the 5′ region of the gray short-tailed opossum UCP1 locus (Figure 3). Notably, two separate regions within the platypus read display homology to the opossum UCP1 exon 1 sequence, revealing what appears to be a 186 bp insertion in the platypus exon 1 sequence. The original platypus start codon also appears to be mutated to “AAG” thus translocating the predicted 30 bp ‘exon 1′ of the platypus 176 bp upstream of the gray short-tailed opossum start codon (Figure 3). By contrast, BLAST searches of platypus RNA sequencing projects (SRX182802, SRX17144, SRX17145, SRX081892, SRX081881, SRX081882, SRX328084, SRX328085, SRX081887-SRX081890) reveal an intact UCP1 mRNA sequence (Supplemental File 5) that differs from the predicted coding sequence. Briefly, the platypus mRNA coding sequence indicates that the predicted 30 bp “exon 1” coding sequence is not translated, that there is no insertion in exon 1 of the platypus, and that the ATG start codon found in other mammals is indeed intact at the expected position (i.e., there is a misassembly error in the predicted GenBank sequence).
Figure 3.
Dot plot comparison of the gray short-tailed opossum UCP1 exon 1 vs. a section of the platypus UCP1 gene occurring between TBC1D9 and ELMOD2 (accession number: NW_001794248.1). Sequence alignments of the platypus (top) and gray-short tailed opossum (bottom) are provided with the potential coding sequences indicated in bold; putative splice sites are underlined. Note that two regions within the platypus clearly display homology to the opossum exon 1 (199–226 and 400–520), suggesting the presence of a 186 bp insertion in the platypus exon 1 sequence. The blue shaded area represents the region where an automated predictor program, which created a seven exon UCP1 gene for the platypus, placed a 30 bp “exon 1” in order to obtain an open reading frame free from premature stop codons (accession number: XM_001512650), though this region shares no homology with exon 1 of the opossum. The original platypus start codon also appears to be mutated to AAG (red font), with the predicted platypus “exon 2” occurring 6 bp downstream of the “ATG” start site in the opossum. Note that these differences between the two species likely arise from a misassembly error in the platypus (see text for details).
UCP1 basal promoter
An alignment of the basal UCP1 promoter for representative species is displayed in Figure 4. Notably, the most upstream GCCCCT motif (nucleotides 1–6 of the promoter alignment; Figure 4) described in the rat by Yubero et al. (1994) is not present in any non-murid species. While the CRE-4 consensus sequence (5′-TGAAGGGC-3′) is similar to that described by Kozak et al. (1994) in mice (5′-TGACGCGC-3′), this site does differ substantially in many species (e.g., common shrew [Sorex araneus], human, etc.) and is absent in the gray short-tailed opossum, walrus, cow, and giraffe (Figure 4). The second and third GCCCCT sites, respectively occurring at 242–248 and 308–315 of the alignment, are relatively well conserved (Figure 4). By contrast, the putative CCAAT site in the rat (Bouillaud et al., 1988) is highly variable in other mammals. The TATA box described by Bouillaud et al. (1988) is intact in the majority of species including all marsupials where it occurs as a 5′-TATAARR-3′ sequence 260–280 upstream of the ATG start codon of exon 1. While a 5′-TATAAGG-3′ sequence is found ~200 bp upstream of the platypus UCP1 coding sequence, the validity of this site is uncertain due to a misassembly in this region of the GenBank sequence (see above). Interestingly, the walrus motif contains a T → A mutation causing a 5′-TAAATAA-3′ sequence, while the panda, white rhinoceros, horse, and bats share a 5′-TACAWAA-3′ sequence. Among species that possess pseudogenized UCP1 genes, an intact TATA box still remains ~290 bp upstream of the African elephant (L. africana) and manatee (T. manatus) coding sequence while the closely related Cape rock hyrax (Procavia capensis) deviates from the consensus (5′-TACGTGA-3′). Similarly, the pig retains a TATA box identical to that of the cow, camel, and giraffe (5′-GATATAA-3′), though a number of mutations in cetaceans have resulted in a sequence (5′-GACGTCAA-3′) that is virtually unrecognizable as a TATA box (Figure 4).
Figure 4.
UCP1 basal promoter elements alignment for select mammalian species with putative protein binding motifs indicated. Highlighted sites indicate shared nucleotides to the species in which the motif was first described (mouse or rat) and the typical TATA box (5′-TATAAAA-3′) sequence (Xu et al., 1991). The consensus sequence represents the simple majority based on species for which the UCP1 gene is intact. Species with documented UCP1 pseudogenes (Gaudry et al., 2017) are denoted in red font and were not included in the consensus calculations.
CpG island
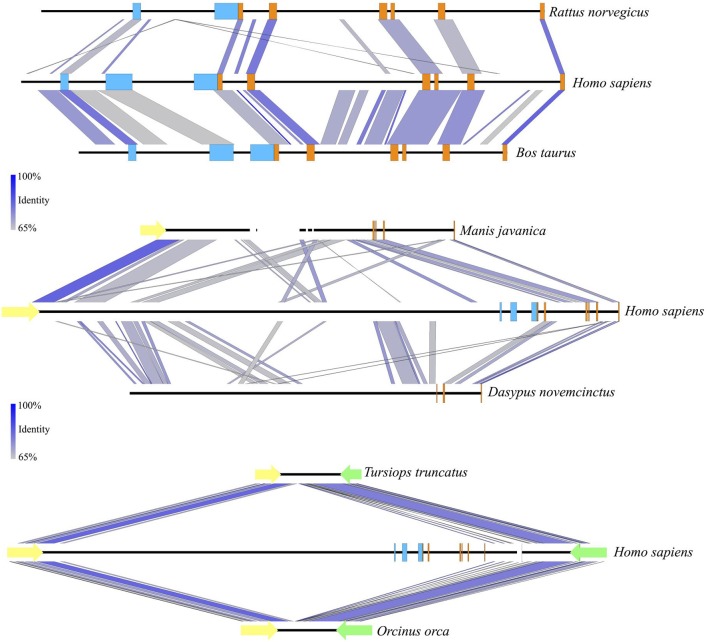
CpG islands meeting the criteria of Gardiner-Garden and Frommer (1987) were not detected in the monotreme or marsupial assemblies. Conversely, a CpG island within or immediately upstream of exon 1 was identified in 91 of 113 eutherian species with available sequence coverage for this region (Table 1). The presence of the CpG island was found to vary extensively among small-bodied species as it was detected in the common shrew, but is absent from the European hedgehog (Erinaceus europaeus) and star-nosed mole (Condylura cristata; Table 1). Many rodent species (e.g., mouse, rat), known to express functional BAT, also lack a CpG island (Table 1). Similarly, among the four afroinsectiphilians examined, a CpG island was only identified in the lesser hedgehog tenrec (containing 39 CpG dinucleotides), despite a relatively high number of CpG sites (37–41) located between 600 bp upstream and 200 bp downstream of the start codon in the other three species. Conversely, CpG islands were identified in closely related paenungulates (elephants, sirenians, and hyraxes), which have >50 CpG dinucleotides in the same region, and armadillos—despite both of these groups having a non-functional UCP1. Among artiodactyls, CpG islands were detected in camels, the okapi (Okapia johnstoni), and all whale UCP1 pseudogenes (except for the killer whale and bottlenose dolphin for which the entire gene is deleted; Figure 5), but not the giraffe or the pig (Sus scrofa). This element is also missing in the pangolin pseudogenes, which is likely due to deletion of a portion of the gene upstream of exon 3 (Figure 5).
Figure 5.
Sequence identity comparisons of the UCP1 genes of the rat, cow, pangolin, armadillo, bottlenose dolphin, and killer whale vs. the human. All DNA sequences are shown 5′ (left) to 3′ (right). UCP1 exons 1–6 are denoted with orange rectangles while UCP1 upstream transcriptional regulatory elements are denoted in light blue (enhancer box, putative regulatory region, CpG island; from left to right). Gaps in sequence coverage are represented by white rectangles. Notably, the putative regulatory region is absent in the rat, but conserved in the cow. Upstream regulatory elements also appear to have been deleted in the Javan pangolin and armadillo, which have deletions of UCP1 exons 1–2, and 3–5, respectively. Deletion of the entire UCP1 gene between TBC1D9 (yellow arrows) and ELMOD2 (green arrows) has occurred in bottlenose dolphin and killer whale ~8–15 MYA (Gaudry et al., 2017) and included the upstream regulatory elements. Sequence identity percentage is represented with a color scale.
Putative regulatory region (PRR)
A distinct PRR was found to be present in 97 of the 125 eutherian mammals examined for which sequence is available (Table 1), though this element was not observed in the platypus or gray short-tailed opossum (Figure 6). PRRs were observed from all afrotherians, but not the armadillo, a xenarthran (Table 1), though insertions within this region are prevalent in the elephant shrew, lesser hedgehog tenrec, and aardvark (Figure 6). By contrast, the dot plots of the elephant and manatee—for which UCP1 is pseudogenized—reveal a high conservation of the PRR with virtually no indels, though only the 3′ half of the PRR is present in the hyrax (Figure 6). As seen for the cow (Figure 5), giraffe, camel, and several whales (Figure 6), the PRR is conserved among most artiodactyls, but is missing in the pig UCP1 pseudogene (Figure 6) and deleted in the bottlenose dolphin, killer whale, and Javan pangolin (Figure 5). A PRR is also absent in several species known to express functional BAT, including the shrew and star-nosed mole, several bats (Myotis spp. and Eptesicus fuscus, etc.), and many rodents (Table 1), including the mouse and rat (Figures 5, 6). Similarly, both Canis familiaris and Lycaon pictus lack a PRR, despite this feature being present in all other carnivores (Table 1). The transcription factor binding sites identified within PRRs of selected species using rVista 2.0 are listed in Table 2. PPAR, DR1, DR3, DR4, CREB, and CEBP sites are relatively common within this region in species with and without a functional UCP1 locus.
Figure 6.
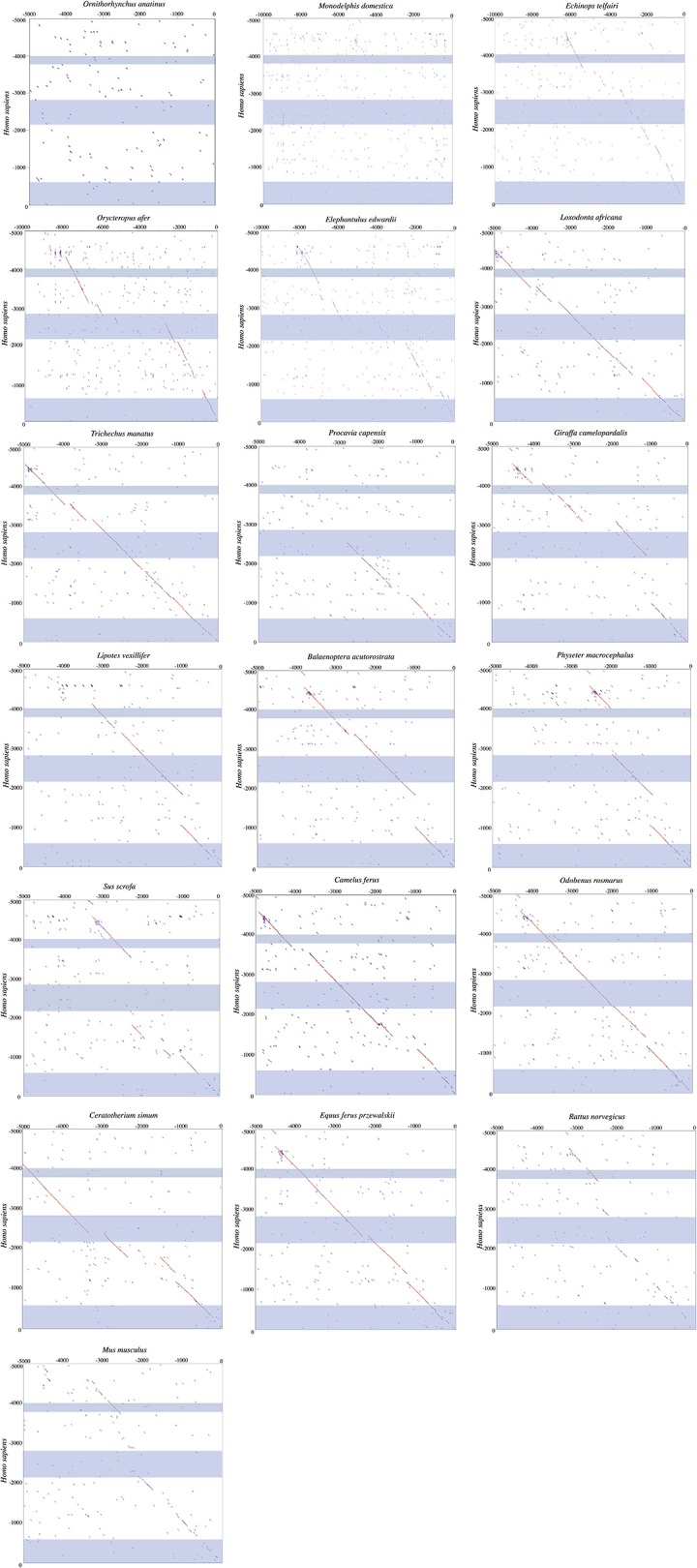
Dot plots of the 5,000 or 10,000 bp upstream of UCP1 exon 1 of select mammalian species compared to the upstream sequence of humans. Blue shading represents the UCP1 enhancer (~ −4,000 to −3,800 in human), putative regulatory region (~ −2,700 to −2,500 in human), and promoter/CpG island (−600 to 0 in human), in that order, from top to bottom.
UCP1 enhancer
UCP1 enhancer sequences were retrieved for 121 eutherian species (Table 1). Enhancer boxes were typically found within 5 kb upstream of exon 1, however, for some members of the afroinsectiphilia (i.e., aardvark and elephant shrew), the enhancer occurs at ~ –7.5 kb (Figure 6). Dot plots of the upstream regions of the platypus and the gray short-tailed opossum reveal no evidence for a UCP1 enhancer (Figure 6), suggesting it is absent within both monotremes and marsupials.
Contrary to the findings of Shore et al. (2012), who noted the absence of an enhancer in the upstream region of the common marmoset (Callithrix jacchus), American pika (Ochotona princeps), thirteen-lined ground squirrel (Spermophilus tridecemlineatus), common shrew, and European hedgehog, we identified this element in each of these species except the hedgehog. The contig encompassing hedgehog UCP1 CDS (accession number: AMUD01193160.1), however, only extends 1126 bp upstream of exon 1 and BLAST searches failed to provide hits of a UCP1 enhancer located on other contigs, thus its presence or absence from the genome remains inconclusive. Similarly, low sequencing coverage likely explains the apparent lack of a UCP1 enhancer in the zebu (Bos indicus), Brazilian guinea pig (Cavia apera), and desert woodrat (Neotoma lepida), as enhancers have been recovered from their close phylogenetic relatives (Table 1).
The enhancer is highly conserved in large-bodied species with intact UCP1 loci (i.e., rhinoceroses, camels, giraffe, and pinnipeds) as well as several species with UCP1 pseudogenes (e.g., elephantids, sirenians, suids, equids, and some cetaceans; Table 1). However, seven species lack both a UCP1 enhancer and an intact UCP1. For instance, the entire UCP1 gene including the enhancer has been deleted in the killer whale and bottlenose dolphin (Figure 5). The enhancer has also been deleted in the sperm whale (Physeter macrocephalus; Figure 6), yet it remains present in the baiji (Lipotes vexillifer) and all baleen whales, indicating an independent loss in both the sperm whale and delphinids. The dot plots also fail to provide evidence for an UCP1 enhancer in the Cape rock hyrax, though this element is present in other paenungulates for which this gene is also pseudogenized (Figure 6). Sequence identity comparisons also suggest the enhancer is lost in pangolins and the nine-banded armadillo (Figure 5 and Table 1). Interestingly, BLAST searches failed to identify this regulator in the WGS contigs or SRA of the two-toed sloth (Choleopus hoffmanni), although partial coverage was recovered for the extinct giant ground sloth (M. darwinii) from a pair of SRA reads (Figure 7).
Figure 7.
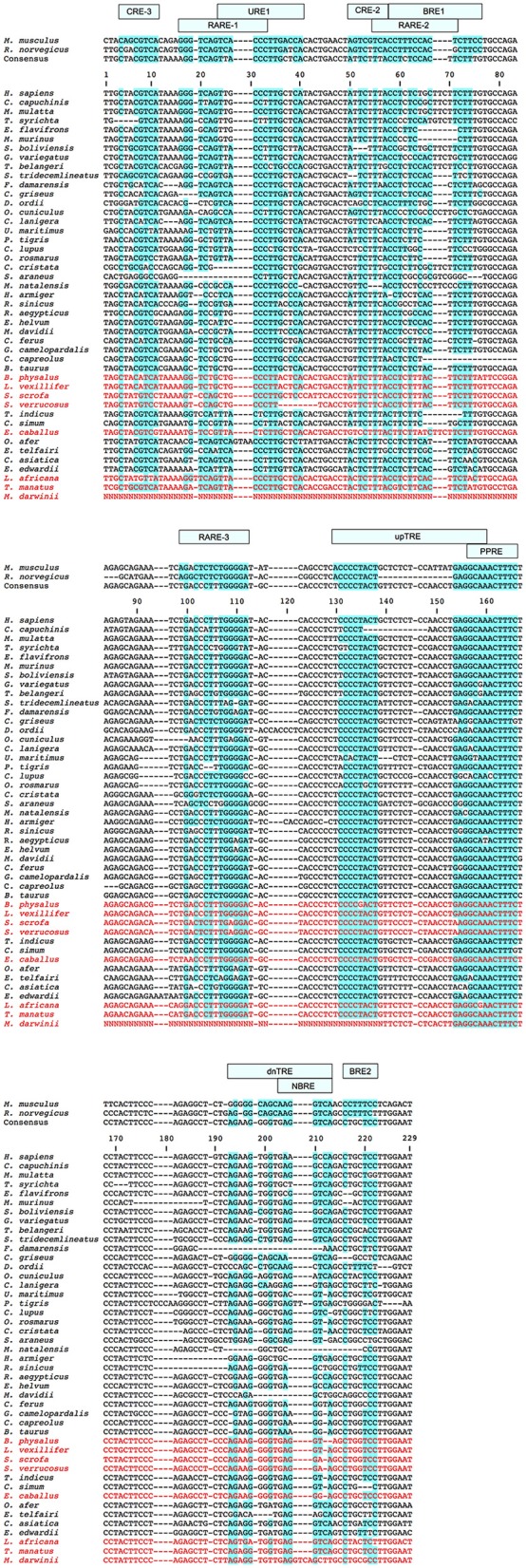
UCP1 enhancer alignment for select eutherian species. Sequences highlighted in blue denote the degree of conservation relative to transcription factor binding sites first described in mice or rats (see also Figure 2). The consensus sequence represents the simple majority based on species for which the UCP1 gene is intact. Species with documented UCP1 pseudogenes (Gaudry et al., 2017) are denoted in red font and were not included in the consensus calculations.
Dot plots of the murid (rat and mouse) upstream sequence (Figure 6) illustrate marked divergence from humans with the exception of a small region encompassing the UCP1 enhancer. By contrast, the upstream sequence of many laurasiatherians, and even paenungulates lacking an intact UCP1 (e.g., elephants and manatees) is surprisingly similar to that of humans (Figure 6). In fact, pairwise sequence comparisons of these enhancers vs. that of the human reveal that this region is more highly conserved (>80%) in large-bodied species that both possess and lack an intact UCP1 than the mouse (74%) and rat (69%) UCP1 (data not show), despite the latter sharing a more recent common ancestor with humans. This pattern is mirrored in the UCP1 gene tree (Figure 8) as many small-bodied lineages (i.e., afroinsectiphlians, myomorph rodents, vesper bats, and most notably, eulipotyphlans) display long branch lengths indicative of high rates of molecular evolution that are comparable to those of many species with UCP1 pseudogenes (e.g., pangolins, pigs, armadillo, and hyrax). Canines are also worth noting, as their branch is highly elongated compared to other carnivores. By contrast, short branches found for most large-bodied species, even among those with non-functional UCP1 (e.g., paenungulates, cetaceans, and equids), reflect low nucleotide substitution rates.
Figure 8.
Maximum likelihood UCP1 coding sequence gene tree illustrating substitution rates in several eutherian lineages (eulipotyphlans, canids, afroinsectiphilians, vesper bats, myomorph rodents; boxed in blue) that are comparable or higher than lineages with UCP1 pseudogenes (denoted in red). Branch lengths represent the number of nucleotide substitutions per site.
Enhancer region alignments revealed a number of marked differences within transcription factor binding motifs among species (Figure 7). For instance, while the CRE-3 site contains a set of core nucleotides (5′-CGTCA-3′) that are highly conserved in most eutherians, mutations to one or two nucleotides within this region are observed in a number of species (e.g., C. cristata, Dipodomys ordi, Cricetulus griseus), while the 5′ portion of this site appears to be deleted in the Philippine tarsier (Tarsius syrichta). Notably, the CRE-3 motif was detected in each species for which the enhancer was screened in rVista except for C. cristata (Table S1). Various mutations to this motif are also found in species with a pseudogenized UCP1 (e.g., elephants, pigs, whales, and horses; Figure 7). The RARE-1 site is especially conserved in the section that overlaps with the URE1 motif, where the consensus sequence (5′-TTACCCTTGCTCA-3′) closely resembles the mouse URE1 site proposed by Sears et al. (1996). However, mutations at sites (e.g., nucleotide positions 32–33 of the alignment in Figure 7) shown to block transcription binding in mice (Sears et al., 1996) are observed in several species with intact UCP1 (e.g., rabbit; Oryctolagus cunculus, Philippine tarsier; T. syrichta, white rhino; C. simum, and tapir; T. indicus). The aardvark displays a 4 bp insertion occurring within the URE1 that results in a single nucleotide (C → A) substitution to this motif. Notably, among species lacking a functional UCP1, the Javan warty pig (S. verrucosus) exhibits a marked disruption to the URE1 site.
The CRE-2 motif is well conserved among most eutherians, however, the consensus eutherian sequence (5′-ATTCTTTA-3′; Figure 7) is a poor match to the mouse 5′-AGTCGTCA-3′ sequence (Kozak et al., 1994). Indeed, of seven species for which the enhancer region was screened using rVista, this site was identified as a cAMP response element only within the mouse (Table S1). Notably, several species with an intact UCP1 display deletions within the CRE-2 motif (e.g., black capped squirrel monkey; Simiri boliviensis, thirteen-lined ground squirrel; S. tridecemlineatus, and natal long-fingered bat; Miniopterus natalensis). Similarly, the two TTCC motifs described for the mouse BRE-1 site (Kozak et al., 1994) are not found in any non-murid eutherians. This region, however, is TC-rich in nearly all species with a single convergent TTCC site found in the dog and natal long-fingered bat (Figure 7). In contrast, the AT-richness of the BRE-1/RARE-2 region is substantially increased in horses, whales, and pigs—all of which lack a functional UCP1—relative to species with an intact gene.
The RARE-3 site consensus sequence (5′-TGACCCTTTGGGGAT-3′; Figure 7) is strongly conserved among eutherians with the exception of a 2-bp deletion in the tiger (Panthera tigris). The PPRE motif predicted by Jastroch et al. (2008) is also a highly conserved element within the UCP1 enhancer, with a consensus sequence of 5′-GCAAACTTTC-3′. Of note, a PPARG (or PPARγ) site with a consensus sequence of 5′-CAAACTTTCTCCTACTT-3′ was identified to overlap with this PPRE motif in six of the seven species (all except for the mouse) for which the enhancer was screened using rVista (Table S1). Conversely, the rat upTRE motif (Rabelo et al., 1995) appears to have arisen from a 14 bp deletion in this species, and is therefore not present in other lineages (Figure 7). Additionally, the white-headed capuchin (Cebus capuchinis) and polar bear (Ursus maritimus), both of which likely express functional BAT, have deletions within the putative upTRE region. The 5′ portion of the dnTRE motif (5′-AGGGCAGCAAGGTCA-3′) described by Rabelo et al. (1995) is also exclusive to the rat, as the consensus sequence (5′-AGAAGGGGTGAGGTCA-3′) has numerous differences and an insertion [bold]; deletions to this region are also found in the Damaraland mole-rat (Fukomys damarensis), Myotis spp. bats, and the lesser hedgehog tenrec (Figure 7). The NBRE site, which overlaps with the 3′ region of the dnTRE, is not strongly conserved in all species, with nucleotide deletions in artiodactyls, the Damaraland mole-rat, great roundleaf bat (Hipposideros armiger), David's myotis, and natal long-fingered bat, and insertions in both the tiger and the giant ground sloth (Figure 7). The most crucial nucleotides of the BRE-2 motif (5′-TTCC-3′; bases 219–222 of the enhancer alignment; Figure 7) described by Kozak et al. (1994) are only found in mice (the species in which it was first described) and the Chinese rufous horseshoe bat (Rhinolophus sinicus).
Discussion
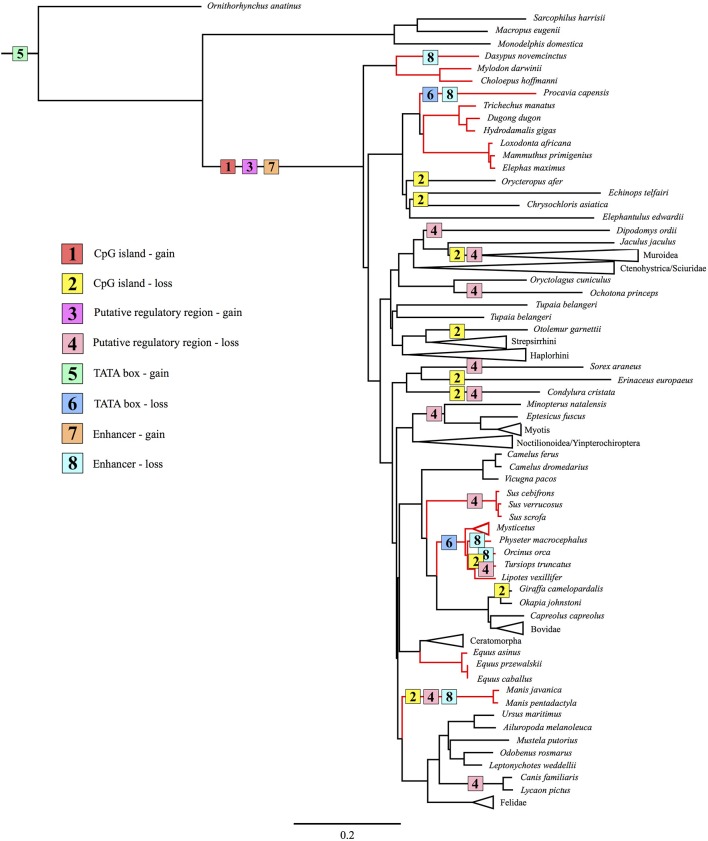
No traces of an enhancer, PRR, or CpG island were detected in the upstream region of the platypus or gray short-tailed opossum loci, though both appear to possess a TATA box within the proximal promoter. By contrast, each of these elements were observed in afrotherians, euarchontoglirans, and laurasiatherians, while a portion of the UCP1 enhancer was also obtained in a single xenarthran, the giant ground sloth, a species that went extinct during the late Pleistocene ~12,000 years ago (Moore, 1978). We can thus deduce that the UCP1 gene of stem mammals contained a TATA box, while the other transcriptional regulatory elements evolved in a common ancestor of eutherians as proposed by Jastroch et al. (2008). However, despite functioning as a hypothetical methylation site (CpG island) or encompassing putative transcription factor binding sites in some species (PRR), these motifs are not required for BAT transcription, as exemplified by high UCP1 expression within the BAT of mice and rats (Pedersen et al., 2001; Wu et al., 2012), which lack both of these elements. Indeed, these elements have repeatedly been lost in eutherian mammals (Figure 9). Shore et al. (2012) reached a similar conclusion as roughly half of the eutherian species they examined lacked a PRR and a CpG island. Given the proposed function of the CpG island as a regulator of UCP1 tissue-specific expression (Kiskinis et al., 2007), a lower level of methylation in BAT as opposed to other tissues would be expected, however, Shore et al. (2012) discovered that the UCP1 CpG island remains virtually un-methylated in BAT, white adipose tissue, and liver despite greatly reduced UCP1 expression levels in the latter two tissues. Therefore, the function of this region remains unclear, however, Shore et al. (2012) did characterize a CpG island in the zebrafish suggesting its presence could be an ancestral condition of the UCP1 gene that was lost in non-eutherian mammals, but retained (and again lost) in some eutherians (see Figure 9).
Figure 9.
Maximum likelihood species tree based on 41 gene segments (50,911 base pairs) composed of both coding and non-coding regions illustrating the gain and loss of known UCP1 regulatory elements (CpG island, PRR, TATA box, enhancer) through the evolutionary history of Mammalia. Red branches indicate lineages with a non-functional UCP1 gene (Gaudry et al., 2017).
Alignment of the proximal promoter CRE-4 site among representative eutherians reveals that the 5′-TGACGCGC-3′ sequence proposed by Kozak et al. (1994) is conserved in the rat, but deviates considerably in the shrew, cow, and human, which are known to express functional BAT (Heaton, 1972; Alexander et al., 1975; Przełecka, 1981). Thus, while the CRE-4 site may play an important role within the murid lineage, it likely does not apply to other eutherians. Similarly, the CCAAT box proposed by Bouillaud et al. (1988) in the rat is highly variable among eutherians (and even among rodents), thus is also unlikely to be a key site for promoter activity. Of the three GCCCCT sites proposed by Yubero et al. (1994), only the two located proximal to exon 1 are conserved, however, to our knowledge transcription factors that bind to these nucleotides have not yet been identified. Overall, the TATA box of the UCP1 promoter is highly conserved in most eutherians, but does vary in some species. For instance, the shared TACA box variant among the horse, rhino, bats, and panda is interesting given that bats and bears possess discernible BAT (Rowlatt et al., 1971; Thomas et al., 1990). While TATA box variants of the flowering plant Arabidopsis thaliana, including the 5′-TACAAAAG-3′ sequence, can still bind the TATA binding protein (TBP) without any structural modifications to the protein, transcription activity levels are substantially (76–85%) lower compared to the 5′-TATAAAAG-3′ sequence (Patikoglou et al., 1999). Considering the high level of TBP conservation among eukaryotes (Peterson et al., 1990), its ability to bind TATA box variants may also apply to mammals. The same T → C transition at the third nucleotide position has been described in the TATA (TACA) box of rabbit uteroglobin with respect to the rat and human, causing a 7-fold reduction in activity when binding to TBP (Klug et al., 1994). However, two other proteins (TATA core factor and TATA palindrome factor) present in uteroglobin-expressing cells bind the TACA box with high efficiency to promote cell specific-expression of the protein (Klug et al., 1994), thus the same possibility may apply to bears, bats, and rhinos. The mutated 5′-TAAATAA-3′ site of the walrus retains a high A/T richness and can thus likely still efficiently bind the TBP (Patikoglou et al., 1999). Notably, the TATA boxes of the hyrax and cetacean UCP1 pseudogenes are poorly conserved, likely due to mutations accumulating under neutral evolution (Figure 9).
In general, the UCP1 enhancer appears to be among the most crucial elements of transcriptional regulation as it is one of the few highly conserved regions in the upstream sequence between humans and rodents (Figure 6). Indeed, excluding four species with low sequence coverage (see below), the enhancer was recovered from all eutherians with an intact UCP1 gene, and therefore is likely essential for UCP1 expression in BAT. This conclusion is at odds with that of Shore et al. (2012), who incorrectly deduced that this region was deleted in a number of species. While we were unable to retrieve an enhancer in four species (i.e., European hedgehog, zebu, Brazilian guinea pig, and desert woodrat), contigs of these species either do not extend ~5 kb upstream of UCP1 exon 1 or contain large sequencing gaps.
In concert with our prediction that large body size may be associated with relaxed selection pressures for UCP1 expression, several anomalies among putative transcription factor binding motifs exist that could be indicative of degradation of these elements were observed. For instance, rhinoceroses display a deletion within the BRE-2 site, and multiple mutations occur within the dnTRE and NBRE regions of camels and the alpaca (Vicugna pacos). However, deletions also occur within these regions of some small-bodied species (Damaraland mole-rat, lesser hedgehog tenrec, and Myotis spp. bats) that also have an intact UCP1, while felids display a highly divergent nucleotide sequence within this 3′ region of the enhancer box. Overall, it thus seems unlikely that these transcriptional regulatory element mutations would substantively impact UCP1 expression in the large-bodied species. Notably, UCP1 regulatory regions (enhancer, PRR, CpG island, promoter) are also present in all large-bodied species (e.g., rhinoceroses, pinnipeds, camel), except the giraffe where a CpG island was not detected (Table 1). Again, this finding suggests that the UCP1 protein may be present in BAT and/or beige tissue of these lineages, highlighting the need for future investigation of UCP1 expression in these species.
In support of our hypothesis that transcriptional regulators would be deteriorated or lost in eutherians with UCP1 pseudogenes, at least five independent lineages (sperm whale, hyrax, pangolins, armadillo, and the family delphinidae [killer whale and bottlenose dolphin]) lack an UCP1 enhancer (Figure 9); notably the TATA box is also lost/mutated in these lineages. By contrast, we identified several lineages (elephantids, sirenians, suids, equids, and some cetaceans) that retain a highly conserved enhancer despite inactivation of their UCP1 genes >20 MYA (Gaudry et al., 2017). The presence of a conserved enhancer upstream of the pig UCP1 pseudogene was also noted by Shore et al. (2012), who suggested that an added function might explain its high degree of sequence identity to that of humans. One such added function could be pleiotropy; the regulation multiple genes (He and Zhang, 2006). Indeed, evolutionary constraint increases (i.e., a higher degree of purifying selection) in mammalian enhancers with increasing pleiotropy (Hiller et al., 2012). Considering that pleiotropic enhancers are not uncommon among mammals (Hiller et al., 2012), this hypothesis cannot be entirely discounted. However, the loss of an UCP1 enhancer in the sperm whale, killer whale, bottlenose dolphin, hyrax, armadillo, and pangolins implies that this enhancer is non-pleiotropic. The apparent conservation of most enhancer elements in the other species for which UCP1 is pseudogenized (e.g., baleen whales, elephants, sirenians, horses) is presumably in part due to an inherently slow rate of molecular evolution arising from their large body size. Indeed, other pseudogenized genes (e.g., AMBN, AMEL, ENAM, and MMP20) in baleen whales and the Steller's sea cow (H. gigas) show exceptionally low rates of molecular decay (Meredith et al., 2011; Springer et al., 2015). Consequently the high (>80%) enhancer sequence identity shared between UCP1-pseudogenized species (horse, minke whale, pig, baiji, bowhead whale, African elephant, and manatee) and humans is not surprising. It thus also remains possible that slow rates of DNA evolution may explain the retention and conservation of these regulatory elements in some large-bodied species with intact UCP1 CDS. By contrast, the higher sequence divergence in rats and mice, which share only 69 and 74% of UCP1 enhancer similarity with humans, respectively, can likely be attributed to a relatively fast mutation rate.
Surprisingly, an elevated mutation rate is also evident in the UCP1 coding sequence of canids as well as the small-bodied lesser hedgehog tenrec, myomorph rodents, vesper bats, and, particularly within members of the order eulipotyphla (Figure 8). While selection pressure analyses indicate that the UCP1 coding sequences of these species display relatively low dN/dS ratios (<0.22; Gaudry et al., 2017), associated with functional conservation of the protein, the very high substitution rates in these groups equate to a substantively elevated number of non-synonymous amino acid substitutions relative to other eutherian lineages (Figure S1). Notably, these high substitution rates are not found for UCP2 or UCP3 sequences of these species (cf. Figure 1), suggesting that this is not solely a size-dependent phenomenon. Consequently these lineages provide intriguing comparative opportunities to study functional UCP1 attributes, as BAT-mediated NST is likely crucial for thermoregulation in these lineages.
A key finding of this study is that several transcription factor binding motifs first described in either mice or rats (BRE-1, BRE-2, upTRE, dnTRE) appear to be restricted to this clade of mammals. Other enhancer motifs (URE1, CRE-2, RARE-2, NBRE) presumed to be key for transcription factor binding in murid rodents (Kozak et al., 1994; Rabelo et al., 1996; Sears et al., 1996; Kumar et al., 2008) are also mutated in other eutherian lineages (Figure 7). Although both single point mutations (Bokar et al., 1988) or combination of mutations (Rabelo et al., 1996) have been shown to alter transcription factor binding to some of these motifs in murid rodents, the effect of the observed differences to these motifs in other eutherians needs to be assessed. Nonetheless, the rVista enhancer screening (Table S1) demonstrates that a number of putative transcription factor binding elements (e.g., CRE-2, PPARG) are not shared between murid rodents and the consensus sequence. This analysis also suggests that components of the transcriptional control of UCP1 expression may be differentially regulated among eutherian mammals. For example, the CRE-3 element was identified in each species selected for screening except for the star-nosed mole (Table S1). By contrast, the high level of sequence identity of the PPRE and RARE-3 elements across Placentalia (Figure 7) indicates that their function has remained strongly constrained throughout eutherian evolution, and is suggestive that they are universally required for the regulation and specificity of UCP1 transcription.
Conclusions
To our knowledge, this study represents the broadest comparative analysis of UCP1 transcriptional regulatory elements among mammals. Our results demonstrate that the CpG island and PRR are not universally conserved among BAT-expressing eutherians and thus are likely not required for UCP1 transcription. In contrast, the TATA box and two of the three GCCCCT sites in the promoter are highly conserved and presumably play a transcriptional role, while the CRE-4 and CCAAT sites differ substantially among eutherians and likely are unimportant. While a UCP1 enhancer was found to be present in every eutherian superorder (Xenarthra [partial], Afrotheria, Laurasiatheria, Euarchontoglires), its absence among non-eutherian mammals supports the hypothesis that it originated with the rise of BAT in a stem placental ancestor. Within this region, however, the specificity and importance of the upTRE, dnTRE, URE1, CRE-2, RARE-2, NBRE, BRE-1, and BRE-2 enhancer elements first described from rats and mice are uncertain as these motifs differ substantially—but generally remain highly conserved—in other BAT-expressing eutherians. Conversely, the RARE-3 and PPRE motifs are among the most highly conserved putative transcription factor binding elements and are likely functional across the eutherian phylogeny. Finally, while some UCP1-less species still retain a UCP1 enhancer, this sequence conservation is presumably due to a slow rate of neutral evolution. Nonetheless, lack of an enhancer in seven UCP1-less species strongly suggests this element is non-pleiotropic.
Author contributions
MG conceived of the project, designed research, prepared DNA libraries, performed hybridization capture experiments, conducted sequencing and genome-mining, performed comparative bioinformatic analyses, prepared the figures, interpreted the results, and drafted the manuscript. KC conceived of the project, designed research, interpreted the results, and revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Eske Willerslev, Rasmus Havmøller, and Tom Gilbert for providing DNA/tissue samples and Mark Springer for providing DNA samples and sequence data for the 41-gene species tree. This study was funded by a National Sciences and Engineering Research Council (NSERC) Discovery Grant (238838) and an NSERC Discovery Accelerator Supplement (412336) to KC.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00670/full#supplementary-material
References
- Alexander G., Bennett J. W., Gemmell R. T. (1975). Brown adipose tissue in the new-born calf (Bos taurus). J. Physiol. 244, 223–234. 10.1113/jphysiol.1975.sp010793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg F., Gustafson U., Andersson L. (2006). The uncoupling protein 1 gene (UCP1) is disrupted in the pig lineage: a genetic explanation for poor thermoregulation in piglets. PLoS Genet. 2:e129. 10.1371/journal.pgen.0020129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A. C., Silva J. E. (1987). Optimal response of key enzymes and uncoupling protein to cold inbrown adipose tissue depends on local T3 generation. Am. J. Physiol. 253, E255–E263. [DOI] [PubMed] [Google Scholar]
- Bokar J. A., Roesler W. J., Vandenbark G. R., Kaetzel D. M., Hanson R. W., Nilson J. H. (1988). Characterization of the cAMP responsive elements from the genes for the alpha-subunit of glycoprotein hormones and phosphoenolpyruvate carboxykinase (GTP). Conserved features of nuclear protein binding between tissues and species. J. Biol. Chem. 263, 19740–19747. [PubMed] [Google Scholar]
- Bouillaud F., Raimbault S., Ricquier D. (1988). The gene for rat uncoupling protein: complete sequence, structure of primary transcript and evolutionary relationship between exons. Biochem. Biophys. Res. Commun. 157, 783–792. 10.1016/S0006-291X(88)80318-8 [DOI] [PubMed] [Google Scholar]
- Brand M. D., Esteves T. C. (2005). Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2, 85–93. 10.1016/j.cmet.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Brent G. A., Moore D. D., Larsen R. P. (1991). Thyroid hormone regulation of gene expression. Annu. Rev. Physiol. 53, 17–35. 10.1146/annurev.ph.53.030191.000313 [DOI] [PubMed] [Google Scholar]
- Cannon B., Nedergaard J. (2004). Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359. 10.1152/physrev.00015.2003 [DOI] [PubMed] [Google Scholar]
- Echtay K. (2007). Mitochondrial uncoupling proteins-What is their physiological role? Free Radic. Biol. Med. 43, 1351–1371. 10.1016/j.freeradbiomed.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 19, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. (1987). CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261–282. 10.1016/0022-2836(87)90689-9 [DOI] [PubMed] [Google Scholar]
- Gaudry M. J., Jastroch M., Treberg J. R., Hofreiter M., Paijmans J. L. A., Starret J., et al. (2017). Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci. Adv. 3:e1602878. 10.1126/sciadv.1602878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M., Seale P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263. 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- He X., Zhang J. (2006). Toward a molecular understanding of pleiotropy. Genetics 173, 1885–1891. 10.1534/genetics.106.060269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton J. M. (1972). The distribution of brown adipose tissue in the human. J. Anat. 112, 35–39. [PMC free article] [PubMed] [Google Scholar]
- Heldmaier G. (1971). Nonshivering thermogenesis and body size in mammals. J. Comp. Physiol. 73, 222–248. [Google Scholar]
- Herpin P., Damon M., Le Dividich J. (2002). Development of thermoregulation and neonatal survival in pigs. Livest. Prod. Sci. 78, 25–45. 10.1016/S0301-6226(02)00183-5 [DOI] [Google Scholar]
- Hiller M., Schaar B. T., Bejerano G. (2012). Hundreds of conserved non-coding genomic regions are independently lost in mammals. Nucleic Acids Res. 40, 11463–11476. 10.1093/nar/gks905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. A., Jastroch M., Stoneking M., Klingenspor M. (2009). Molecular evolution of UCP1 and the evolutionary history of mammalian non-shivering thermogenesis. BMC Evol. Biol. 9:4. 10.1186/1471-2148-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastroch M., Buckingham J. A., Helwig M., Klingenspor M., Brand M. D. (2007). Functional characterization of UCP1 in the common carp: uncoupling activity in liver mitochondria and cold-induced expression in the brain. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 177, 743–752. 10.1007/s00360-007-0171-6 [DOI] [PubMed] [Google Scholar]
- Jastroch M., Withers K. W., Taudien S., Frappell P. B., Helwig M., Fromme T., et al. (2008). Marsupial uncoupling protein 1 sheds light on the evolution of mammalian nonshivering thermogenesis. Physiol. Genomics 32, 161–169. 10.1152/physiolgenomics.00183.2007 [DOI] [PubMed] [Google Scholar]
- Kiskinis E., Hallberg M., Christian M., Olofsson M., Dilworth S. M., White R., et al. (2007). RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J. 26, 4831–4840. 10.1038/sj.emboj.7601908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenspor M., Fromme T. (2012). Brown adipose tissue, in Adipose Tissue Biology, ed Symonds M. E. (New York, NY: Springer; ), 39–79. [Google Scholar]
- Klingenspor M., Fromme T., Hughes D. A., Manzke L., Polymeropoulos E., Riemann T., et al. (2008). An ancient look at UCP1. Biochim. Biophys. Acta 1777, 637–641. 10.1016/j.bbabio.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Klug J., Knapp S., Castro I., Beato M. (1994). Two distinct factors bind to the rabbit uteroglobin TATA-box region and are required for efficient transcription. Mol. Cell. Biol. 14, 6208–6218. 10.1128/MCB.14.9.6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak U. C., Kopecky J., Teisinger J., Enerback S., Boyer B., Kozak L. P. (1994). An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol. Cell. Biol. 14, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Liu D., Wang H., Robidoux J., Collins S. (2008). Orphan nuclear receptor NOR-1 enhances 3′,5′-cyclic adenosine 5′-monophosphate-dependent uncoupling protein-1 gene transcription. Mol. Endocrinol. 22, 1057–1064. 10.1210/me.2007-0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Cao C., Tao C., Ye R., Dong M., Zheng Q., et al. (2017). Cold adaptation in pigs depends on UCP3 in beige adipocytes. J. Mol. Cell. Biol. [Epub ahead of print]. 10.1093/jmcb/mjx018 [DOI] [PubMed] [Google Scholar]
- Loots G., Ovcharenko I. (2004). rVista 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32, W217–W221. 10.1093/nar/gkh383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux R. J., Harper M. E. (2011). Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic. Biol. Med. 51, 1106–1115. 10.1016/j.freeradbiomed.2011.06.022 [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. 10.1016/S0378-1119(99)00368-6 [DOI] [PubMed] [Google Scholar]
- McGaugh S., Schwartz T. S. (2017). Here and there, but not everywhere: repeated loss of uncoupling protein 1 in amniotes. Biol. Lett. 13:20160749 10.1098/rsbl.2016.0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab B. K. (1983). Energetics, body size, and the limits to endothermy. J. Zool. 199, 1–29. 10.1111/j.1469-7998.1983.tb06114.x [DOI] [Google Scholar]
- Meredith R. W., Gatesy J., Cheng J., Springer M. S. (2011). Pseudogenization of the tooth gene enamelysin (MMP20) in the common ancestor of baleen whales. Proc. Biol. Soc. 278, 993–1002. 10.1098/rspb.2010.1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Kircher M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 6:Pbdprot5448. 10.1101/pdb.prot5448 [DOI] [PubMed] [Google Scholar]
- Moore D. I. (1978). Post-glacial vegetation in the South Patagonian territory of the giant ground sloth, Mylodon. Bot. J. Linean. Soc. 77, 177–202. 10.1111/j.1095-8339.1978.tb01398.x [DOI] [Google Scholar]
- Mzilikazi N., Jastroch M., Meyer C. W., Klingenspor M. (2007). The molecular and biochemical basis of nonshivering thermogenesis in an African endemic mammal, Elephantulus myurus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R2120–R2127. 10.1152/ajpregu.00427.2007 [DOI] [PubMed] [Google Scholar]
- Nakajima N., Horikoshi M., Roeder R. G. (1988). Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol. Cell. Biol. 8, 4028–4040. 10.1128/MCB.8.10.4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkrug R., Goetze N., Exner C., Lee Y., Ganjam G. K., Kutschke M., et al. (2013). Brown fat in a protoendothermic mammal fuels eutherian evolution. Nat. Commun. 4:2140. 10.1038/ncomms3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkrug R., Polymeropoulos E. T., Jastroch M. (2015). Brown adipose tissue: physiological function and evolutionary significance. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 185, 587–606. 10.1007/s00360-015-0907-7 [DOI] [PubMed] [Google Scholar]
- Patikoglou G. A., Kim J. L., Sun L., Yang S. H., Kodadek T., Burley S. K. (1999). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13, 3217–3230. 10.1101/gad.13.24.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson L. E., Liwanag H. E., Hammill M. O., Burns J. M. (2014). To each its own: thermoregulatory strategy varies among neonatal polar phocids. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 178, 59–67. 10.1016/j.cbpa.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Pedersen S. B., Bruun J. M., Kristensen K., Richelsen B. (2001). Regulation of UCP1, UCP2, and UCP3 mRNA expression in brown adipose tissue, white adipose tissue, and skeletal muscle in rats by estrogen. Biochem. Biophys. Res. Commun. 288, 191–197. 10.1006/bbrc.2001.5763 [DOI] [PubMed] [Google Scholar]
- Peterson M. G., Tanese N., Pugh B. F., Tjian R. (1990). Functional domains and upstream activation properties of cloned human TATA binding protein. Science 248, 1625–1630. 10.1126/science.249.4971.844-b [DOI] [PubMed] [Google Scholar]
- Polymeropoulos E. T., Jastroch M., Frappell P. B. (2012). Absence of adaptive nonshivering thermogenesis in a marsupial, the fat-tailed dunnart (Sminthopsis crassicaudata). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 182, 393–401. 10.1007/s00360-011-0623-x [DOI] [PubMed] [Google Scholar]
- Przełecka A. (1981). Seasonal changes in ultrastructure of brown adipose tissue in the common shrew (Sorex araneus L). Cell Tissue Res. 214, 623–632. 10.1007/BF00233501 [DOI] [PubMed] [Google Scholar]
- Rabelo R., Reyes C., Schifman A., Silva J. E. (1996). A complex retinoic acid response element in the uncoupling protein gene defines a novel role for retinoids in thermogenesis. Endocrinology 137, 3488–3496. [DOI] [PubMed] [Google Scholar]
- Rabelo R., Schifman A., Rubio A., Sheng X., Silva J. E. (1995). Delineation of thyroid hormone-responsive sequences within a critical enhancer in the rat uncoupling protein gene. Endocrinology 136, 1003–1013. 10.1210/endo.136.3.7867554 [DOI] [PubMed] [Google Scholar]
- Rowlatt U., Mrosovsky N., English A. (1971). A comparative survey of brown fat in the neck and axilla of mammals at birth. Biol. Neonate 17, 53–83. 10.1159/000240303 [DOI] [PubMed] [Google Scholar]
- Saito S., Saito C. T., Shingai R. (2008). Adaptive evolution of the uncoupling protein 1 gene contributed to the acquisition of novel nonshivering thermogenesis in ancestral eutherian mammals. Gene 408, 37–44. 10.1016/j.gene.2007.10.018 [DOI] [PubMed] [Google Scholar]
- Sears I. B., MacGinnitie M. A., Kovacs L. G., Graves R. A. (1996). Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol. Cell. Biol. 16, 3410–3419. 10.1128/MCB.16.7.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore A., Emes R. D., Wessely F., Kemp P., Cillo C., D'Armiento M., et al. (2012). A comparative approach to understanding tissue-specific expression of uncoupling protein 1 expression in adipose tissue. Front. Genet. 3:304. 10.3389/fgene.2012.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore A., Karamitri A., Kemp P., Speakman J. R., Lomax M. A. (2010). Role of UCP1 enhancer methylation and chromatin remodeling in the control of UCP1 expression in murine adipose tissue. Diabetologia 51, 1164–1173. 10.1007/s00125-010-1701-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Signore A. V., Paijmans J. L., Vélez-Juarbe J., Domning D. P., Bauer C. E., et al. (2015). Interordinal gene capture, the phylogenetic position of Steller's sea cow based on molecular and morphological data, and the macroevolutionary history of Sirenia. Mol. Phylogenet. Evol. 91, 178–193. 10.1016/j.ympev.2015.05.022 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. W., Dorais M., Bergeron J. M. (1990). Winter energy budgets and cost of arousals for hibernating little brown bats, Myotis lucifugus. J. Mamm. 71, 475–479. 10.2307/1381967 [DOI] [Google Scholar]
- Umesono K., Murakami K. K., Thompson C. C., Evans R. M. (1991). Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65, 1255–1266. 10.1016/0092-8674(91)90020-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya F., Peyrou M., Giralt M. (2017). Transcriptional regulation of the uncoupling protein-1 gene. Biochimie 134, 86–92. 10.1016/j.biochi.2016.09.017 [DOI] [PubMed] [Google Scholar]
- Wu J., Boström P., Sparks L. M., Ye L., Choi J. H., Giang A. H., et al. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Thali M., Schaffner W. (1991). Upstream box/TATA box order is the major determinant of the direction of transcription. Nucleic Acids Res. 19, 6699–6704. 10.1093/nar/19.24.6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubero P., Vinas O., Iglesias R., Mampel T., Villarroya F., Giralt M. (1994). Identification of tissue-specific protein binding domains in the 5'-proximal regulatory region of the rat mitochondrial brown fat uncoupling protein gene. Biochem. Biophys. Res. Commun. 204, 867–873. 10.1006/bbrc.1994.2540 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.