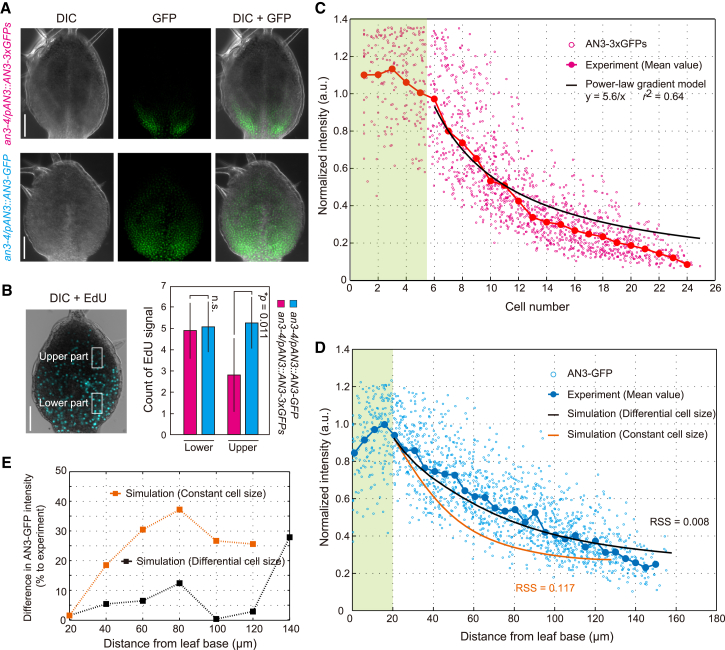
Figure 5.
AN3 gradient formation by pure diffusion process in a growing leaf tissue. (A) Leaf primordia from an3-4/pAN3::AN3-3xGFPs (upper) and an3-4/pAN3::AN3-GFP (lower). DIC (left), GFP fluorescence (middle), and their merged images (right) are shown. Scale bars, 50 μm. (B) Distribution of proliferating cells in leaf primordia. A DIC image merged with EdU-positive signal (blue) is shown (left). Numbers of EdU-positive nuclei per 20 × 40 μm area (white rectangles) at leaf lower and upper parts were counted. The mean ± SD with the p value from the Student’s t-test are shown (n > 12). n.s., non-significant (p > 0.05). Scale bars, 50 μm. (C and D) Distribution profiles for AN3-3xGFPs and AN3-GFP fluorescence intensities along the leaf proximal-to-distal axis. Normalized intensities of AN3-3xGFPs (C) and AN3-GFP (D) in nuclei are plotted as a function of cell number and distance from the leaf base, respectively. The mean of AN3-3xGFPs and AN3-GFP intensities binned by cell position (red dots in C) and distance from the leaf base at 5-μm intervals (blue dots in D) are indicated. A fitting curve with a power-law gradient to the AN3-3xGFPs profile in the targeted tissue is shown in black in (C). Simulated curves through the 4-μm constant-cell-size field (orange) and the differential (from 4 to 8 μm) cell-size field (black) are shown in (D). a.u., arbitrary unit. The green shaded area indicates the region of source tissue. RSS, residual sum of squares. (E) Difference in AN3-GFP intensity between simulated and experimentally determined profiles. The differences of the simulated curve with 4-μm constant cell size (orange) and with differential cell size (black) against experimentally measured values are plotted as a function of the distance from the leaf base. Mean deviations were calculated based on the data for 20–120 μm from the leaf base. All parameters used are summarized in Table S2. To see this figure in color, go online.