Fig. 5.
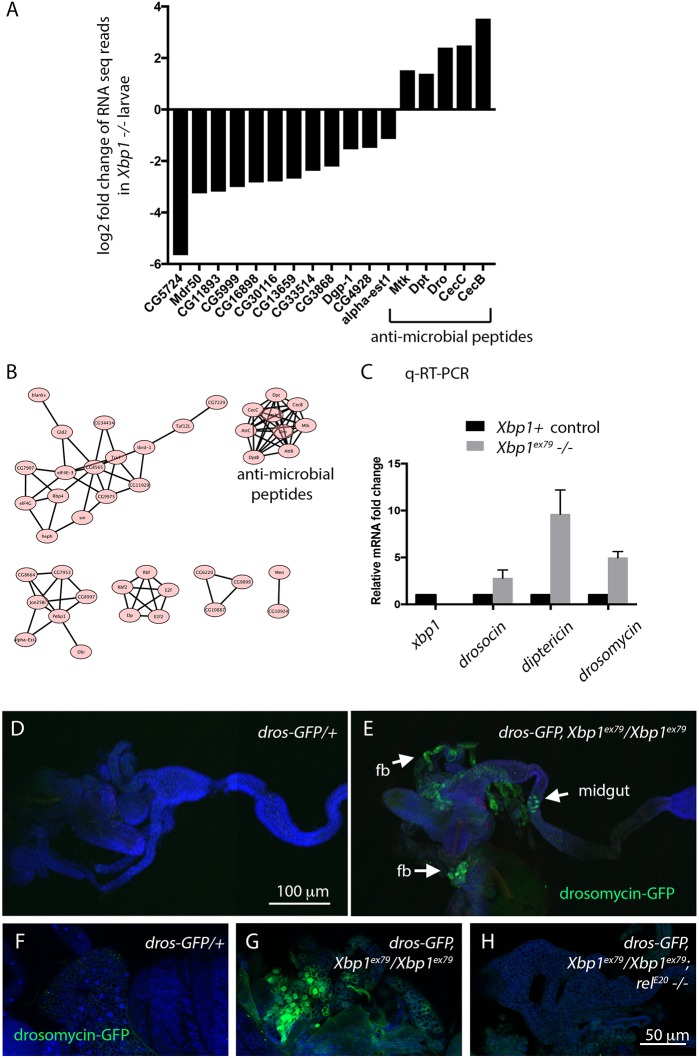
The innate immune response is activated in Xbp1 mutant larvae. (A) A select set of genes with altered gene expression (adjusted P value <0.05) in Xbp1 mutant larvae. The full list is in Table S1. The y-axis shows the log2 fold change of RNAseq reads in Xbp1 mutants. Genes shown here are those previously reported to be tunicamycin-inducible genes in the fly (reported in Chow et al., 2013; left 12 bars), and those encoding anti-microbial peptides (right five bars). (B) Network analysis of the genes that are induced in Xbp1 mutants. Those that encode anti-microbial peptides form a major cluster. (C) Validation of AMP transcript induction through qRT-PCR. qRT-PCR against Xbp1 was performed to confirm that the samples were from Xbp1ex79−/− null larvae. As representative AMPs, drosocin, diptericin and drosomycin transcripts were examined. (D,E) drosomycin-GFP, a reporter for an anti-microbial peptide expression (green) in wild-type (D) and Xbp1 mutant (E) second-instar larvae. TO-PRO-3 (blue) was used to show the outline of tissues. D and E are composite images. The scale bar for D and E is shown in D. (F–H) Induction of drosomycin-GFP in the fat body of Xbp1 mutants (wild-type in F and Xbp1−/− in G) is abolished in the mutant background of rel20 (H), which encodes a Rel family of immune responsive transcription factor. The scale bar for F–H is shown in H.