Figure 6.
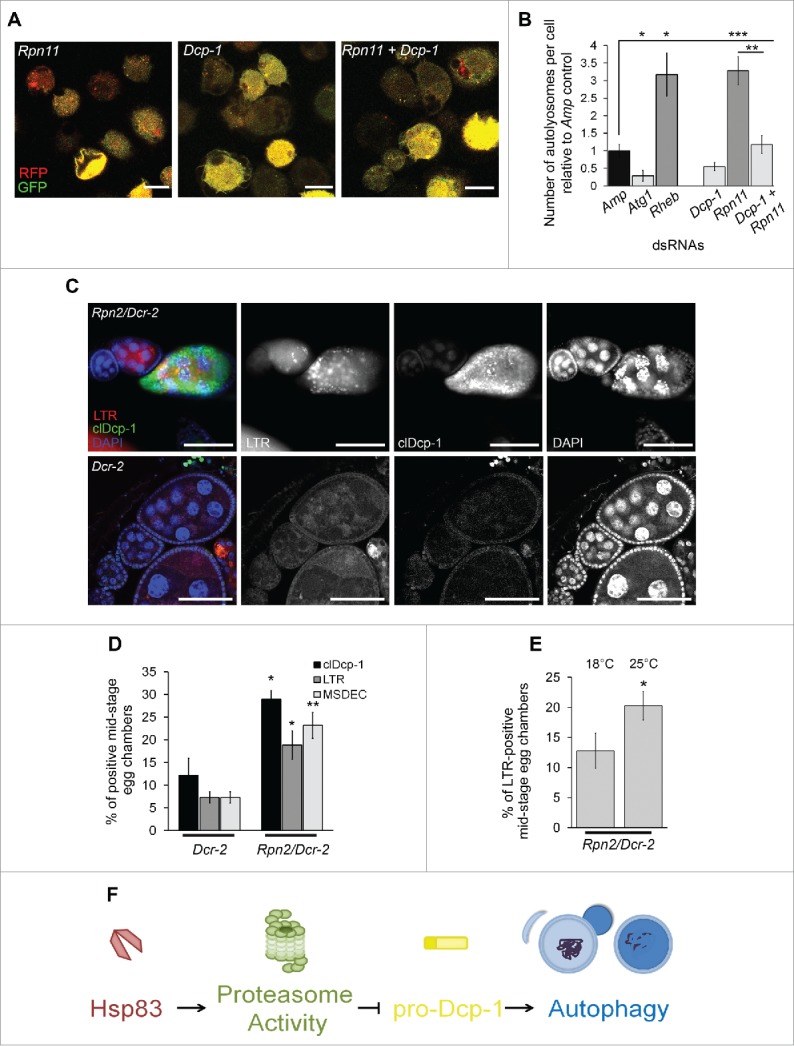
Proteasomal subunit loss results in Dcp-1-dependent compensatory autophagy. (A, B) The number of autolysosomes per cell was quantified in S2 cells stably expressing GFP-RFP-Atg8a and treated with the indicated dsRNAs. (A) Representative images of GFP-RFP-Atg8a S2 cells following treatment with the indicated dsRNAs; scale bars: 10 μm. (B) All counts were normalized to the Amp dsRNA control. Atg1 and Rheb dsRNA's served as controls for decreasing and increasing the number of autolysosomes, respectively. Error bars represent ± SEM and statistical significance was determined using one-way ANOVA with a Dunnet post-test, *P < 0.05, **P < 0.01,***P < 0.001 (C, D) Females were collected from flies with the UAS maternal driver kept at 25°C with the genotypes UAspDcr-2/+; nosGAL4/+ (Dcr-2) and Rpn2-RNAI/UAspDcr-2;nosGAL4/+ (Rpn2/Dcr-2). (C) Representative images of MSECs from Rpn2/Dcr-2 and Dcr-2 kept at 25°C. Ovaries were imaged with clDcp-1 antibody, LTR and DAPI; scale bars: 50 μm. (D) MSECS from Dcr-2 and Rpn2/Dcr-2 flies were scored for cleaved Dcp-1 (clDcp-1), LTR and DAPI. The graph represents the average percentage of MSECs that scored positive for LTR, MSDEC or clDcp-1. Experiments were performed with at least 8 females per genotype (n = 3). Error bars represent ± SEM and statistical significance was determined using the 2-way Student t test, *P < 0.05, **P < 0.01 (E) MSECS in Rpn2/Dcr-2 flies were scored for LTR positivity at 18°C and 25°C. At least 50 MSECS were analyzed per temperature (n = 4). Error bars represent ± SEM and statistical significance was determined using the 2-way Student t test, *P < 0.05 (F) A proposed pathway indicating that Hsp83 functions in basal conditions to contribute to proteasomal activity which suppresses pro-Dcp-1 levels and thus prevents activation of autophagy or cell death.
