ABSTRACT
Decreased luminal endoplasmic reticulum (ER) Ca2+ concentration triggers oligomerization and clustering of the ER Ca2+ sensor STIM1 to promote its association with plasma membrane Orai1 Ca2+ channels leading to increased Ca2+ influx. A key step in STIM1 activation is the release of its SOAR domain from an intramolecular clamp formed with the STIM1 first coiled-coil (CC1) region. Using a truncated STIM1(1–343) molecule that captures or releases the isolated SOAR domain depending on luminal ER Ca2+ concentrations, we analyzed the early molecular events that control the intramolecular clamp formed between the CC1 and SOAR domains. We found that STIM1 forms constitutive dimers, and its CC1 domain can bind the SOAR domain of another STIM1 molecule in trans. Artificial oligomerization failed to liberate the SOAR domain or activate STIM1 unless the luminal Ca2+-sensing domains were removed. We propose that the release of SOAR from its CC1 interaction is controlled by changes in the orientation of the two CC1 domains in STIM1 dimers. Ca2+ unbinding in the STIM1 luminal domains initiates the conformational change allowing SOAR domain liberation and clustering, leading to Orai1 channel activation.
KEY WORDS: STIM1, ORAI1, Ca2+ entry, Endoplasmic reticulum, Ca2+
Highlighted Article: The mechanism by which STIM1 molecules transmit information on changes in ER Ca2+ concentration involves a conformational change in the CC1 domain of the cytoplasmic segment of dimeric STIM1 molecules rather than oligomerization.
INTRODUCTION
One of the most intriguing lines of discoveries in the Ca2+ signaling field in the last 10 years has been the identification of the molecules responsible for the major form of store-operated Ca2+ entry (SOCE) pathway (Liou et al., 2005; Roos et al., 2005). SOCE was postulated 30 years ago when it was recognized that lowering the endoplasmic reticulum (ER) Ca2+ content is sufficient to activate a Ca2+ influx pathway (hence the name store-operated) that was distinct from the then known voltage-gated Ca2+ entry mechanisms (Putney, 1986). The identity of the molecules responsible for SOCE had eluded identification for a long time in spite of intense efforts (Parekh and Putney, 2005). Only with the aid of RNAi technology was it possible to finally identify both the Ca2+-sensing unit located in the ER, namely stromal interaction molecule 1 (STIM1) (Liou et al., 2005; Roos et al., 2005), and the Ca2+ channel partner in the plasma membrane, called Orai1 (Feske et al., 2006) or CRACM1 (Vig et al., 2006). An impressive rapid progress in understanding the molecular details of how these two proteins work together followed these seminal discoveries. It was understood that decreased ER luminal Ca2+ and the resultant unbinding of Ca2+ from the luminally located EF-hands of the STIM1 molecule triggers a sequence of events that leads to clustering of the molecules in ER–plasma membrane (PM) junctions where an interaction takes place with the Orai1 Ca2+ channels causing their opening (reviewed in Deng et al., 2009; Derler et al., 2012; Hogan et al., 2010; Soboloff et al., 2012; Zhou et al., 2017). It was also shown that the STIM1 cytoplasmic tail (CT) , which has a single membrane-spanning domain, is sufficient to activate Orai1 channels (Huang et al., 2006), which was soon followed by the identification of the minimum Orai1 activation sequence within the STIM1 CT-domain, called SOAR (Yuan et al., 2009) or CAD (Park et al., 2009).
The striking difference between the potent ability of the SOAR domain and the moderate effect of the full STIM1-CT in activating Orai1 raised the possibility that part(s) of STIM1-CT exerts inhibition on SOAR, and indeed artificial clustering of STIM1-CT at the ER–PM junction was found to increase its potency for Orai1 activation to the level observed with the isolated SOAR domain (Korzeniowski et al., 2010). These observations led to the idea that an intramolecular interaction between the SOAR domain and other parts of the STIM1-CT keeps SOAR in an inactive state, and that this auto-inhibition is relieved upon STIM1 activation and clustering (Korzeniowski et al., 2010). It was then found that the SOAR domain is constitutively liberated when an acidic region (Korzeniowski et al., 2010) or key hydrophobic residues (Muik et al., 2011) are mutated within the coiled-coil domain-1 (CC1) causing a large conformational change of the full STIM1-CT from a closed to an extended conformation (Muik et al., 2011). Because the SOAR domain contains a basic stretch of residues required for Orai1 activation (Calloway et al., 2010; Korzeniowski et al., 2010), initially it was assumed that the acidic region in CC1 and the basic residues in SOAR form an electrostatic clamp (Korzeniowski et al., 2010). Other studies emphasized the role of hydrophobic coiled-coiled interactions (Muik et al., 2011). Recent advances in obtaining structural information (Cui et al., 2013; Stathopulos et al., 2013; Yang et al., 2012) and elegant analysis of the conformational changes upon opening the intramolecular clamp (Zhou et al., 2013) disproved the electrostatic interaction model, and suggested that a conformational change induced within the dimeric CT domain precedes and triggers oligomerization/clustering and the ultimate activation of Orai1 (Fahrner et al., 2014; Ma et al., 2015; Zhou et al., 2013).
In the present study, we examined the early activation process of the STIM1 molecule, namely the conformational requirements of the liberation of the SOAR domain from its autoinhibitory interaction with the STIM1 CC1 domain in the natural ER environment of the molecule. Using a combination of approaches that rely upon the use of intact cells, we show that co-expression of the SOAR domain with STIM1 mutants truncated within the CC1 domain results in a dynamic association between the two molecules that is regulated by the ER Ca2+ store content. The dynamics of this process can be followed in live intact cells allowing us to investigate the earliest events in STIM1 activation. Our data suggest that a conformational switch within a STIM1 dimer rather than oligomerization is the earliest triggering event leading to the liberation of the SOAR domain and to the subsequent molecular events including clustering and activation of the Orai1 channels.
RESULTS
Live-cell imaging reveals a reversible intramolecular interaction between the STIM1 CC1 and SOAR domains
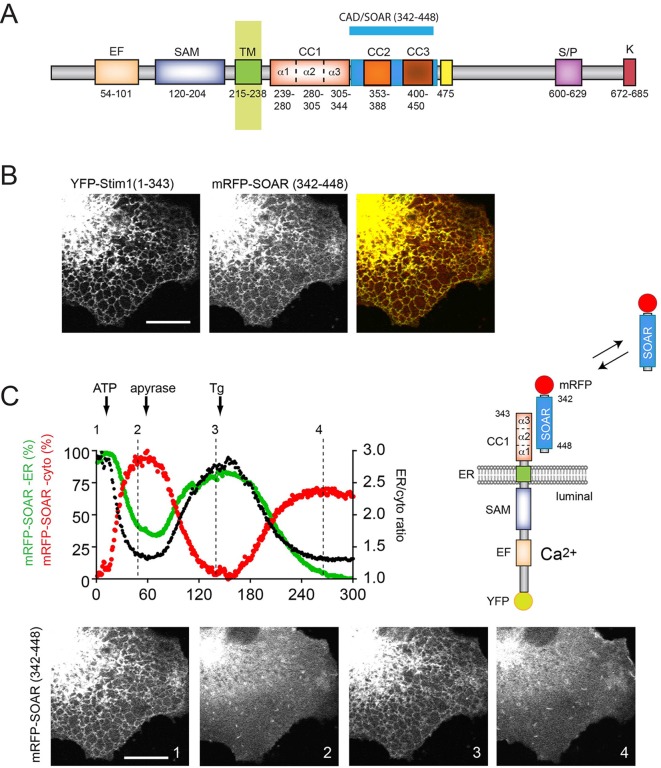
Recent studies examined the conformational requirement for CC1–SOAR interactions in vitro, using recombinant purified proteins (Zhou et al., 2013) or in situ ‘in the cell’ Förster resonance energy transfer (FRET) approaches (Fahrner et al., 2014; Ma et al., 2015). In order to visualize this interaction in situ in the natural ER membrane environment, we generated a series of STIM1 truncations and expressed them together with the SOAR domain (Fig. 1A) for confocal microscopy analysis. Expression of a luminally YFP-tagged STIM1 construct truncated at the end of the CC1 domain [YFP–STIM1(1–343)] together with an mRFP-tagged SOAR domain (342–448) in COS-7 cells showed that the truncated STIM1 was able to anchor the SOAR domain to the surface of the ER (Fig. 1B) (note in the YFP–STIM1 constructs, the YFP is inserted between residues 23 and 24 of STIM1, but for simplicity this is not indicated in the abbreviations). Remarkably, this association was interrupted by depletion of the ER Ca2+ stores by the combined administration of ATP and thapsigargin (Tg) (50 µM and 200 nM, respectively) releasing the SOAR domain from the ER surface to the cytosol. Furthermore, when ATP was added alone and its effect was terminated by the addition of apyrase, the process was reversed, and the SOAR domain re-associated with the ER (Fig. 1C).
Fig. 1.
Dynamic imaging of the interaction between STIM1–CC1 and SOAR domains in situ. (A) Schematics of STIM1 structure with segment boundaries relevant to the present study. Numbering corresponds to the human STIM1 protein. EF, Ca2+ -binding EF-hand motif; SAM, sterile alfa motif; TM, transmembrane segment; CC1, CC2 and CC3, coiled-coil domains; SOAR, minimal Orai1 activation region; S/P, proline-serine-threonine-rich segment; K, polybasic domain. (B) Confocal images showing the localization of YFP–STIM1 truncated at the end of the CC1 domain [YFP–STIM1(1–343)] and the soluble mRFP-tagged SOAR domain (mRFP–SOAR) co-expressed in COS-7 cells. Cells were kept in low-Ca2+ conditions during transfection (regular DMEM with 0.1 mM CaCl2) and transferred to modified Krebs–Ringer buffer containing 2 mM Ca2+ medium 5 min prior to the experiment. (C) Reversible association of the SOAR domain with YFP–STIM1(1–343). Depletion of the ER Ca2+ stores by the addition of ATP (50 µM) or Tg (200 nM) releases the SOAR domain from the ER surface to the cytosol. The effect of ATP is reversed by the addition of apyrase (5 units/ml) resulting in store refilling and recapture of SOAR. Red and green traces show fluorescence intensity changes obtained from a representative mRFP–SOAR time-lapse image in region of interest covering the cytosol and the ER (at the perinuclear ER-dense region), respectively. The black trace is the ER:cytosol ratio intensity change. Still images are shown from the same time-lapse experiment and correspond to the indicated time points. The cartoon shows our explanation for these changes. Scale bars: 10 μm.
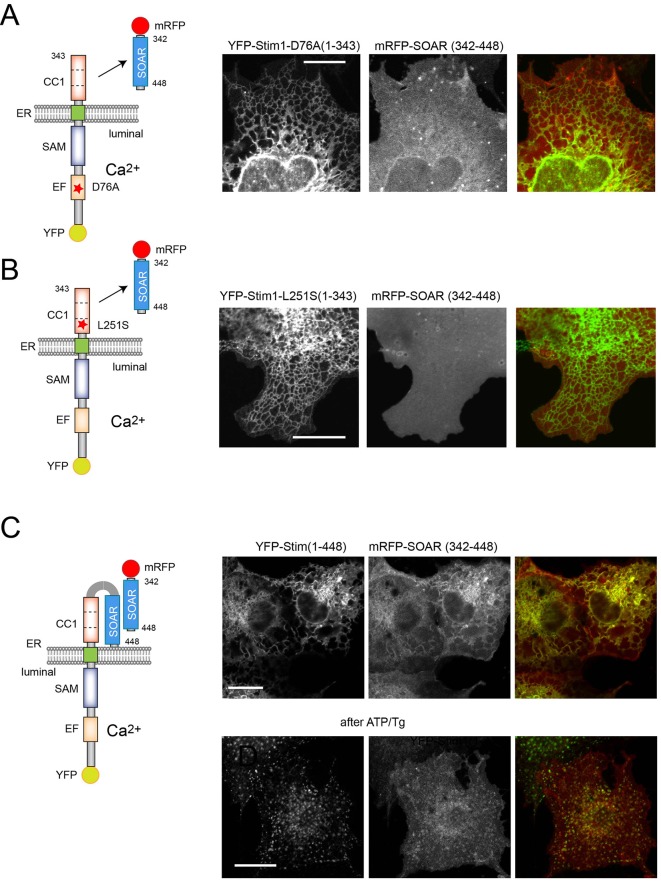
Serial truncations within the CC1 domain showed that YFP–STIM1(1–250) was not able to attract the SOAR domain (Figs S1, S2), but constructs ending at residues 280, 301, 315 and 327 still showed SOAR interaction and responded to ER Ca2+ depletion by releasing the SOAR domain in a reversible manner (Figs S1, S2). This result suggests that the segment 238–280, referred to as CC1α1 (Fahrner et al., 2014), provides the major site of interaction with the SOAR domain. It is important to note that none of the STIM1 constructs listed above showed clustering during this activation process. The only sign of their active state was the release of the SOAR domain from their interaction. Mutations that have been shown to induce the active STIM1 conformation, such as D76A within the luminal Ca2+-sensing domain (Liou et al., 2005) or L251S within the CC1 domain (Muik et al., 2011) also rendered YFP–STIM(1–343) unable to bind the SOAR domain (Fig. 2A,B). Extension of the STIM1 molecule beyond the CC1 region to include the SOAR domain [YFP–STIM1(1–448)] reduced but did not eliminate binding of the expressed soluble SOAR domain. Importantly, this interaction was no longer disrupted by ER Ca2+ depletion, in fact this STIM1(1–448)–SOAR complex showed clustering within the ER upon activation (Fig. 2C; Fig. S2).
Fig. 2.
Activating mutations in STIM1 prevent SOAR docking to CC1. Activating mutations in the YFP–STIM1(1–343) construct abolish the ability of SOAR to bind the CC1 domain. (A) COS-7 cells were transfected with YFP–STIM1(1–343)D76A and mRFP–SOAR. No ER association of the SOAR domain was observed. (B) YFP–STIM1(1–343)L251S also prevents mRFP–SOAR association with the STIM1 mutant. (C) When the SOAR domain is included in the STIM1 sequence [YFP–STIM1(1–448)] the mRFP–SOAR domain still binds to the STIM1 molecule. However, it is not released during Ca2+ store depletion, rather it co-clusters with the STIM1 molecule. We reason that this result reflects a SOAR–SOAR interaction. Here, representative images are shown, but statistical analysis of a large number of cells is shown in Fig. S2. Scale bars: 10 μm.
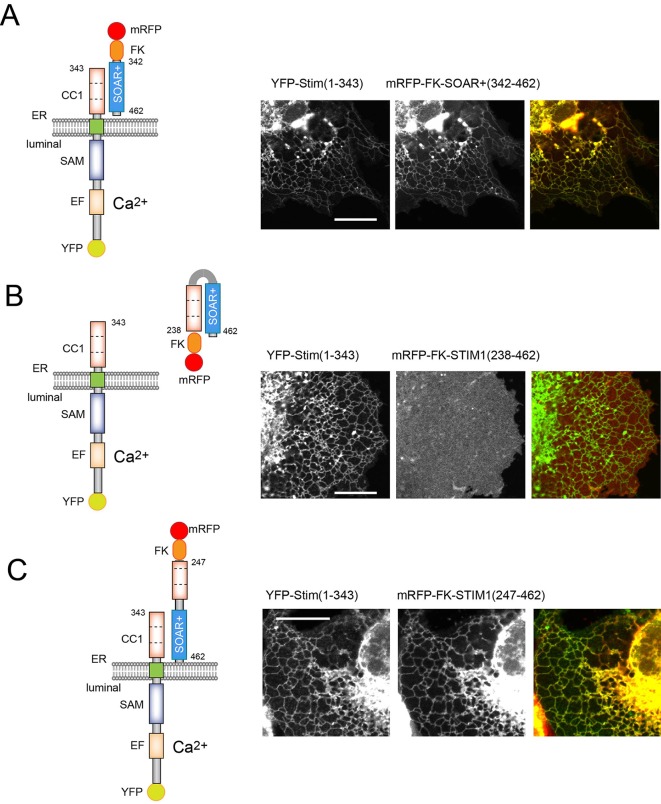
Next, we tested the effect of extending the soluble SOAR domain toward the C-terminus (343–462) (termed as SOAR+) and added an FKBP12 module to its N-terminus (see below for its additional utility). These modifications did not affect the ability of the SOAR domain to interact with YFP–STIM1(1–343) (Fig. 3A) or its response to ATP and Tg (not shown). However, inclusion of the CC1 domain in the soluble SOAR+-containing construct (238–462) prevented its interaction with the YFP–STIM1(1–343) module (Fig. 3B). We interpreted these results as suggestive of the CC1 domain now interacting with the SOAR domain in cis rather than with the CC1 fragment of the YFP–STIM1(1–343) molecule in trans (Fig. 3B). Remarkably, deletion of a small nine-residue fragment from the beginning of the CC1 domain in the CC1–SOAR+ construct (residues 247–462), was sufficient to restore the interaction with YFP–STIM1(1–343), suggesting that the membrane-adjacent residues at the beginning of the CC1 domain are important in stabilizing the conformation of CC1 to be optimal for SOAR binding (Fig. 3C). Notably, however, this small membrane-adjacent segment (between residues 238 and 247) was not sufficient to establish a strong interaction with SOAR since the STIM1 molecule truncated at residue 250 was unable to bind the SOAR domain (Fig. S2).
Fig. 3.
The juxtamembrane region within the CC1 domain is necessary to stabilize the SOAR–CC1 interaction. COS-7 cells were co-transfected with the indicated constructs and imaged by confocal microscopy. (A) A short extension of SOAR and the addition of an FKBP12 module (mRFP–FKBP12–SOAR+; 342–462) does not affect the interaction of SOAR with STIM–CC1 or its response to store depletion (the latter response is not shown here). (B) Inclusion of the CC1 domain in the soluble mRFP–FKBP12–STIM1 construct [mRFP–FKBP12–STIM1(238–462)] prevents its association with the CC1 domain because of the formation of the interaction between the two domains in cis (see cartoon). (C) N-terminal truncation of the CC1 domain in the soluble CC1–SOAR construct restores SOAR interaction with the ER-bound STIM1–CC1 molecule. This indicates that the intramolecular interaction in cis is broken allowing the SOAR–CC1 interaction in trans (see cartoon). Scale bars: 10 μm.
These results were essentially confirmatory and are compatible with those published by the Zhou and Romanin laboratories (Fahrner et al., 2014; Ma et al., 2015) but, importantly, allowed us to monitor the earliest activation events in situ in the cells without expressing Orai1 or having SOAR present on the STIM1 molecule, which, amplifies the activation state by oligomerization.
STIM1 can work as a dimer
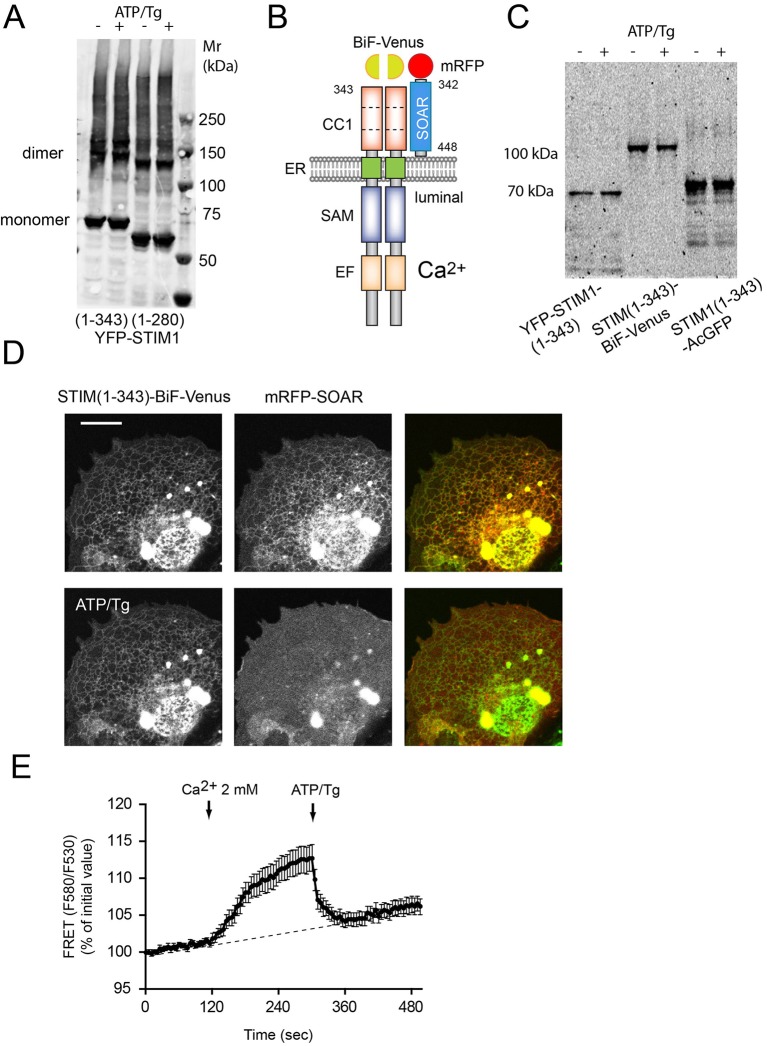
To follow the interaction between the SOAR domain and the STIM1(1–343) construct by FRET analysis, we tagged the STIM1(1–343) construct at its C-terminus with the monomeric Aequorea coerulescens (Ac)GFP. However, this construct was less active in attracting the SOAR domain to the ER than the luminally tagged form (Fig. S3A). This was in contrast to the findings of Ma et al. (Ma et al., 2015) who found that such a C-terminally tagged construct showed a FRET interaction with the SOAR domain. Although, we do not have a good explanation for this discrepancy, our finding might have been caused by interference of the C-terminal AcGFP tag with how the CC1 domain can bind SOAR. Regardless, since our crosslinking experiments suggested that STIM1 molecules exist as dimers even in the resting state (Fig. 4A), we considered the possibility that the CC1 domains work as dimers. To test whether a STIM1(1–343) dimer can bind SOAR, we generated obligate STIM1 dimers using the bimolecular fluorescence complementation (BiF) approach (Kerppola, 2008). Venus(1–155) was fused to the C-terminus of one of the STIM1(1–343) constructs, while Venus(156–end) was fused to the identical STIM1 construct. Expression of the two constructs created fluorescence at the ER, which is only possible if two STIM1 molecules are associated with a single Venus molecule (see cartoon in Fig. 4B). This was proven with native gel electrophoresis that showed fluorescence at the expected molecular size formed from STIM1(1–343) dimers with one fluorescent Venus molecule (Fig. 4C). (It has to be noted that at higher expression levels, prominent YFP signal was localized to the nuclear envelope and generated large ER saccules.) Importantly, however, the SOAR domain was again anchored to the Venus-positive ER structures and responded to ER Ca2+ depletion, as assessed by FRET analysis (Fig. 4D,E). These experiments suggested that the CC1 domains of STIM1 were able to bind the SOAR domain in a dimeric form.
Fig. 4.
C-terminally tagged obligate STIM1-CC1 dimers efficiently dock the SOAR domain. (A) Western blot analysis of cell lysates obtained from COS-7 cells expressing the indicated YFP–STIM1 constructs and cross-linked with 100 µM DSS. Cells in lanes 2 and 4 (+) were stimulated with ATP and Tg (ATP/Tg) for 5 min before crosslinking. STIM1 was detected by using a rabbit anti-STIM1 (N-terminus) antibody and donkey anti-rabbit-IgG secondary antibody (conjugated to IRDye 680LT) and analyzed with an Odyssey imager. Note the dimeric form was present even in quiescent cells. (B) Obligate STIM1–CC1 dimers were formed by using a BiF complementation approach. COS-7 cells were transfected with STIM1(1–343)–BiF–Venus(1–155) and STIM1(1–343)–BiF–Venus(155–end) together with mRFP–SOAR and kept in low-Ca2+ medium for 12–24 h. (C,D) Fluorescence of STIM1 constructs expressed in COS-7 cells, and run in native gels (C) or imaged (D). Note the presence of BiF-complemented STIM1 dimers (D). mRFP–SOAR docks efficiently to dimerized CC1 domains from STIM1(1–343)–BiF–Venus fluorescent constructs. ER Ca2+ store depletion releases the SOAR domain from the STIM–CC1 dimer. (E) Dynamic interaction between the obligate STIM1 CC1 construct and SOAR domain during ER Ca2+ store depletion and refill as assessed by FRET. Energy transfer is detectable when cells previously kept in nominally Ca2+-free medium were treated with 2 mM CaCl2. A FRET decrease is then observed after the addition of ATP/Tg. FRET values are expressed as change relative to initial values. Results are means±s.e.m. (n=30 cells from multiple dishes in two independent experiment).
STIM1 CC1 domains are able to bind SOAR domains in trans
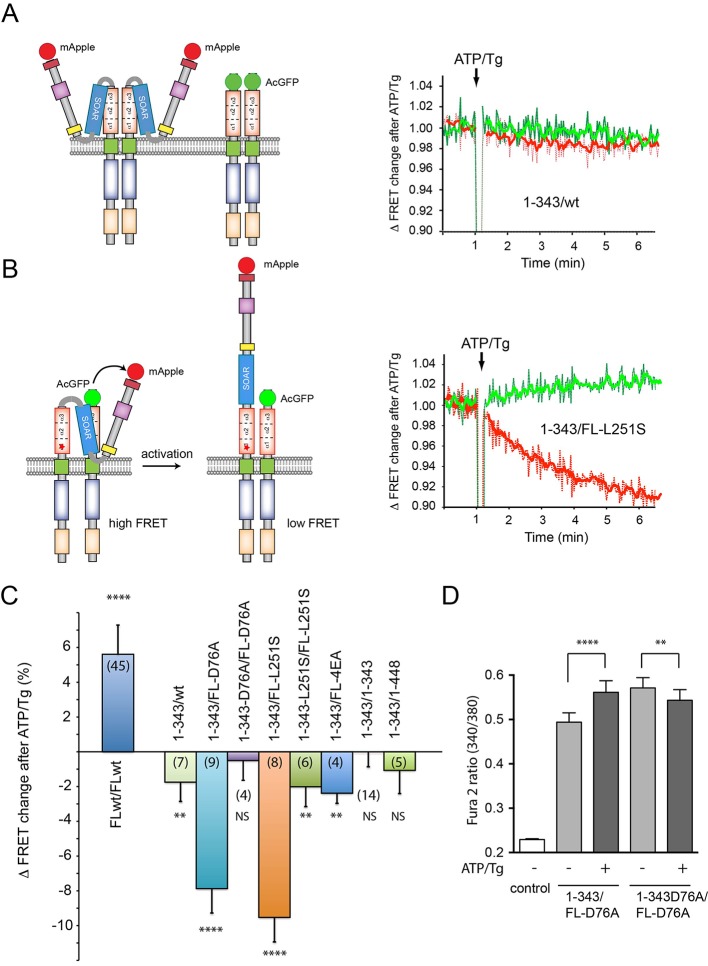
Having found that STIM1 is able to work as a dimer, next we investigated the ability of the STIM1-CC1 domain to bind the SOAR domain when the latter is part of the full-length STIM1 molecule. For this analysis, we tagged full-length STIM1 at the C-terminus with mApple and used the STIM1(1–343) construct tagged with AcGFP at its C-terminus, and expressed the two constructs together for FRET analysis. Although this latter construct was a less optimal binder of cytosolic SOAR (see above), we reasoned that it could still pair with the full-length STIM1. [When both mApple and AcGFP were fused to full-length STIM1, we found an increased FRET, as described before by several laboratories (Covington et al., 2010; Korzeniowski et al., 2016; Liou et al., 2007); the cartoon and original traces are not shown, but FRET change is shown in summary in Fig. 5C.] When we used the full-length STIM1 paired with the 1–343 truncated form, a minor decrease in FRET was observed after ATP and Tg addition (Fig. 5A,C, note that the change in FRET in response to store depletion is plotted on this graph and not absolute FRET values). Remarkably, when the full-length STIM1–mApple was mutated to make it constitutively active (D76A or L251S) and it was co-expressed with the STIM1(1–343)–AcGFP, we observed a massive decrease in the FRET signal upon ER store depletion (Fig. 5B,C). Our explanation for this finding was that the SOAR domain liberated from the CC1 domain of the active (mutated) full-length STIM1, was captured in trans by the CC1 domain of the truncated STIM1 molecule (see cartoon of Fig. 5B). Upon activation, the SOAR domain was then released from the truncated STIM1 CC1 domain causing the decrease in the FRET signal. To determine whether this indeed was the case, we mutated the short STIM1 construct to also make it constitutively active (i.e. unable to bind a SOAR domain). As shown in Fig. 5C, no significant FRET changes were observed after activation when both the full and the truncated STIM1 molecules harbored the activating mutations. These experiments suggested that SOAR domain and CC1 domain interaction could occur in trans, consistent with the idea that STIM1 molecules form constitutive dimers.
Fig. 5.
SOAR domains in STIM1 dimers can interact with the CC1 domain in trans. FRET analysis in COS-7 cell suspension between STIM1 constructs tagged with AcGFP and mApple proteins as donor and acceptor, respectively. (A) No significant FRET change was detected between C-terminally mApple-tagged full-length STIM1 and AcGFP-tagged STIM(1-343) during store depletion. Green traces show donor (AcGFP) fluorescence and red traces show the sensitized acceptor:donor signal ratio. Ratios were normalized to the starting baseline for each recording separately [60 s average before ATP and Tg (ATP/Tg) addition]. Values above one indicate a stimulated positive FRET change and values below one indicate a stimulated decrease in the FRET ratio. Left panel shows cartoon with possible pairing of the two constructs. (B) Significant FRET decreases were detected after ER Ca2+ store depletion in COS-7 cells transfected with the constitutively active version of the full-length STIM1(L251S)-AcGFP as donor and STIM1(1-343)-mApple as the acceptor. The cartoon on left explains how the SOAR domain in the constitutively active mutant STIM1 is free to pair with the CC1 domain of the truncated wild-type STIM1 molecule resulting in a high basal FRET signal that decreases upon STIM1 activation. (C) Summary of FRET changes using different pairing of the full-length and STIM1(1-343) constructs in similar experiments. Note that both activating mutants (D76A or L251S) in the full-length (FL) STIM1 paired with wild-type STIM1(1-343) shows the large FRET decrease upon store Ca2+ depletion. However, pairing the activated full-length STIM1 with the same activating mutations within the STIM1(1-343) partner eliminates the FRET change since now both CC1 domains are in a conformation that is unable to bind the SOAR domain. Curiously, although the 4EA mutation has been found as an activating one in our previous study (Korzeniowski et al., 2010), it has proven to be not so active in this assay. Means±s.e.m. are shown from 3-9 independent experiments (numbers of measurements are indicated in the columns in parentheses) (****P<0.0001; **P<0.01). (D) Cytoplasmic Ca2+ changes after transfection of the indicated STIM1 constructs together with untagged Orai1. Note that the increased basal Ca2+ evoked by the constitutively active full-length STIM1(D76A) is reduced in the presence of the STIM1(1-343) construct but not if the latter also had the D76A mutation. This difference is abolished by ATP/Tg. Means±s.e.m. obtained from 118 and 158 cells in 6 independent experiments for the STIM1(1-343) and STIM1(1-343)D76A groups, respectively (****P<0.0001; **P=0.033).
Competition of the STIM1 CC1 domain for the SOAR domains of constitutively active STIM1 molecules should reduce the activity of the latter in Ca2+ experiments. To test this experimentally, we had to express Orai1 channels, which compete for binding the SOAR domain of the full-length constitutive STIM1 construct and already caused elevated Ca2+. Nevertheless, we found that the STIM1-D76A mutant increased basal Ca2+ to a significantly smaller extent when co-expressed with STIM1(1-343) than when co-expressed with the mutant form of STIM1(1–343)-D76A. This difference disappeared when ATP and Tg was added to the cells (Fig. 5D).
Notably, the 4EA mutation, which was shown to render STIM1 constitutively active in our previous study (Korzeniowski et al., 2010), did not show the pattern that was observed with the other constitutively active STIM1 full-length constructs (Fig. 5C). We assume that the 4EA mutation weakens the intramolecular clamp and only in the presence of Orai1 will render the STIM1 molecule fully active.
Oligomerization only activates STIM1 if it contains the SOAR domain and lacks the luminal domains
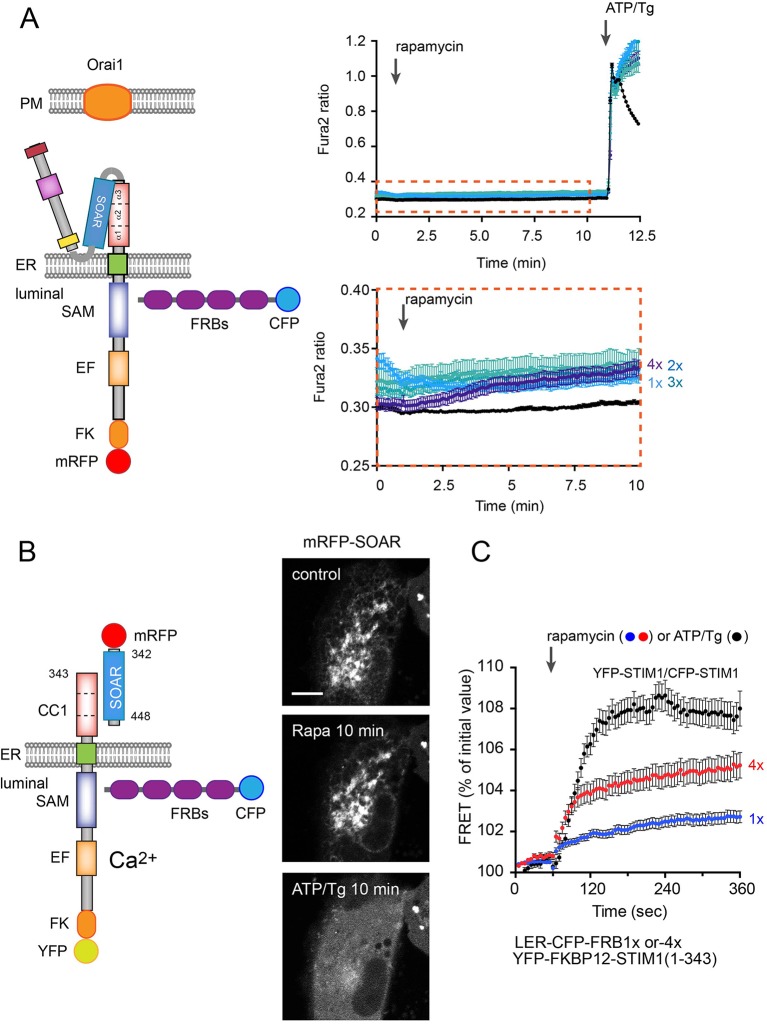
Since current models predict that oligomerization is the earliest event during STIM1 activation, we designed an approach to induce oligomerization of the STIM1 molecules. For this, we generated STIM1 constructs that were tagged at the luminal side with FKBP12 and mRFP. We also created FKBP12–rapamycin-binding (FRB) domain constructs that contained multiple copies of fused FRBs (1×, 2×, 3×, 4×, 5×) and targeted them to the ER lumen. We performed single-cell cytosolic Ca2+ recordings in COS-7 cells. Cells were transfected with the full-length TK-promoter-driven STIM1 luminally tagged with mRFP–FKBP12 and the various ER-lumen-targeted FRBs. We also expressed Orai1 channels to follow the activation process in cytoplasmic Ca2+ measurements. As shown in Fig. 6A, rapamycin-induced oligomerization was a poor activator of the STIM1–Orai1 Ca2+ signaling pathway, although a small activation was measurable using the 4×FRB domains in the ER lumen (see enlarged insert in Fig. 6A). However, ER Ca2+-depletion caused a massive Ca2+ response indicating that the FKBP-tagging did not prevent activation. Similarly, when a YFP–FKBP12–STIM1(1–343) construct was co-transfected with the SOAR domain and the ER-targeted FRB multiples, rapamycin addition failed to release the SOAR domain from the YFP–FKBP12–STIM1(1–343) molecule and only after ER Ca2+ depletion was SOAR released from the ER (Fig. 6B). As shown in Fig. 6C, rapamycin addition did cause increased STIM1 oligomerization when combined with the luminally targeted 4×FRB, suggesting that while oligomerization did happen, it was not able to trigger STIM1 activation.
Fig. 6.
The effect of oligomerization on STIM1 activation. Artificial oligomerization was achieved by expressing ER-lumen-targeted FRB multimers (1× to 4×FRB) together with STIM1 proteins that were also tagged with FKBP12 at the lumen. (A) Single-cell cytosolic Ca2+ recordings were performed in transfected cells loaded with Fura2-AM. Cells were transfected with full-length TK-promoter-driven mRFP–FKBP12–STIM1 (wild type), untagged Orai1 and luminally targeted CFP–FRBs. Rapamycin-induced oligomerization induced a minor elevation of cytosolic Ca2+ only when the 4×FRB was present in the ER (see the bottom trace in A with the enlarged scale). This response was very small compared to the one evoked by subsequent store depletion (ATP/Tg treatment). Results are means±s.e.m. from 133–209 cells recorded in 4–7 separate experiments. (B) Rapamycin-driven oligomerization has no detectable effect on the release of the SOAR domain from a YFP–FKBP12–STIM1(1–343) construct, which is then effectively released by ATP/Tg treatment. Scale bar: 10 μm. (C) FRET measurements between the ER-lumen-targeted FRB multimers and YFP–STIM1. Note that the 4×FRBs cause oligomerization that is significantly higher than those evoked by the 1×FRB and ∼50% of the FRET increase shown by STIM1 molecules upon ATP/Tg treatment. Results are means±s.e.m. from 128, 75 and 43 cells for 1×FRB, 4×FRB and the STIM1–STIM1 groups, respectively, obtained in 6–10 independent experiments.
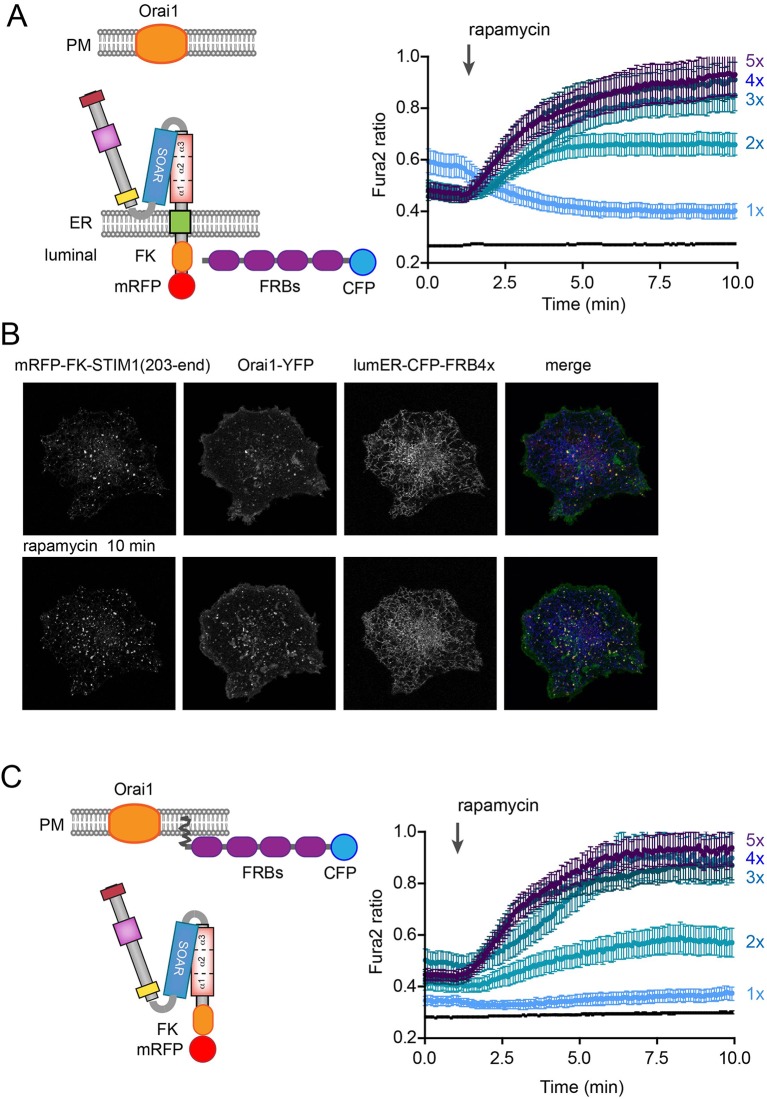
We then further tested the effects of oligomerization on the STIM1 activation process using STIM1 constructs in which the entire luminal segment was replaced by the mRFP–FKBP12 module or in which the entire cytoplasmic STIM1 fragment (without the transmembrane domain) was fused to the mRFP–FKBP12 module (Fig. 7A,B, and 7C, respectively). Again, the FRB multiples (either ER lumen targeted or PM targeted, see cartoon) and Orai1 were co-transfected. Notably, these transfected cells showed a substantially elevated cytoplasmic Ca2+ even before addition of rapamycin. Robust activation was then observed after rapamycin addition with the response increasing with the number of ER-targeted FRB domains reaching a plateau with 4×FRB (Fig. 7A) (curiously the combination with 1×FRB showed a Ca2+ decrease in response to rapamycin). The rapamycin-induced Ca2+ rise was also associated with increased STIM1–Orai1 puncta formation, although puncta were already present before rapamycin addition (Fig. 7B). These results were similar to those reported from the Lewis laboratory using a different oligomerization approach on multimerization-induced ICRAC activation (Luik et al., 2008). We also found similar activation of Ca2+ influx when we recruited cytoplasmic STIM1 molecules to PM-targeted FRB multimers (Fig. 7C). We wanted to test the same manipulations on the ability of the STIM1-CC1 domain to release the SOAR domain. However, we observed that the STIM1 construct lacking the entire luminal domain failed to attract the SOAR domain in most cells. Still, in the few cells (less than 10%) where the SOAR domain was found bound to the STIM1 construct at the ER, rapamycin was without effect (Fig. S3B). These experiments together suggested that STIM1 constructs lacking the luminal domains are already in a partially active state and, in such constructs, oligomerization can effectively drive activation. This, however, requires the presence of the SOAR domain and Orai1, which causes further clustering and helps stabilize an open conformation of the STIM1 molecule.
Fig. 7.
The effect of oligomerization on STIM1 molecules lacking the luminal domains. Artificial oligomerization was achieved by expressing ER-lumen-targeted FRB multimers (1× to 4×FRB) together with STIM1 proteins that were tagged with FKBP12 at the lumen but lacked all luminal domains. (A) COS-7 cell were transfected with TK-driven mRFP–FKBP12–STIM1(203–685) with deleted entire luminal STIM1 domains together with untagged Orai1 and ER-lumen-directed CFP–FRBs. Single-cell cytosolic Ca2+ recordings were performed in transfected cells loaded with Fura2-AM. Cells expressing these constructs already showed higher basal Ca2+ levels, that was further enhanced by rapamycin addition (except for the case of 1×FRB in the ER lumen). Responses were progressively faster and larger as the numbers of FRBs were increased up to 4×FRB. (B) Rapamycin increases clustering of STIM1–Orai1 when oligomerizing with the luminally targeted 4×FRB. Scale bar: 10 μm. (C) Experiments were performed with PM-targeted FRB multimers and a soluble STIM construct also missing the transmembrane domain (see cartoon). Again, the basal Ca2+ was already elevated and further increased by rapamycin, especially with FRB multimers of >3. For the Ca2+ experiments, means±s.e.m. from 46–88 cells are shown recorded in 3–6 separate experiments.
DISCUSSION
Activation of the STIM1–Orai1-mediated Ca2+ influx pathway in ER–PM contact sites is a critically important mechanism in the control of signaling cascades in a variety of cell types and organisms (Hogan and Rao, 2015; Soboloff et al., 2012). The triggering event to engage this mechanism is the decrease in the ER luminal Ca2+ concentration that is sensed by the luminal EF-hands of the single membrane-pass STIM1 molecule (Liou et al., 2005). Ca2+ unbinding on the luminal side then evokes a conformational change in the cytoplasmic segment of the STIM1 molecule that is characterized by an unmasking of the segment called SOAR or CAD that interacts and activates the Orai channels in the PM (Hogan and Rao, 2015). The most obvious sign of this activation cascade is the massive clustering of the STIM1 (and Orai1) molecules in the Ca2+-depleted state of the ER. Recent studies have firmly established that a critical step in STIM1 activation is the liberation of the SOAR domain from an intramolecular clamp that is formed between the SOAR domain and the first membrane-adjacent coiled-coil domain of STIM1, termed CC1 (Fahrner et al., 2014; Korzeniowski et al., 2010; Ma et al., 2015; Muik et al., 2011; Zhou et al., 2013).
The remaining question is how the STIM1 intraluminal domains communicate their conformational change to the cytoplasmic domains across the ER membrane. The prevailing view had been that an initial oligomerization of STIM1 molecules triggers the liberation of the SOAR domain, which then is the seed of further oligomerization and clustering (Covington et al., 2010; Liou et al., 2007). However, recent elegant studies have started to challenge that view. Zhou et al. performed studies with the soluble STIM1 cytoplasmic domain, and suggested that forced dimerization of the molecule at its N-terminal end (i.e. the one just emerging from the ER in the full-length STIM1 molecule) is sufficient to open up the molecule to assume an extended conformation (Zhou et al., 2013). Ma et al. used a FRET approach and found that STIM1(1–343) constructs can attract the SOAR domain and release it upon ER Ca2+ depletion. They suggested that reorientation of the transmembrane domain of STIM1 is the first triggering signal in the activation process. They found that mutation of a key cysteine residue within the transmembrane segment (C227W) is sufficient to activate the STIM1 molecule (Ma et al., 2015). We have attempted to address this question in situ within the cell in the proper ER environment with a readout that does not involve the downstream amplification events driven by the clustering of the SOAR domain. Our approach turned out to be similar to that used by Ma et al. in that we used the association of the SOAR domain with STIM1(1–343) as a readout for monitoring the early activation steps.
Similar to Ma et al. (Ma et al., 2015), we found that a STIM1 molecule truncated at the end of the CC1 domain is capable of attracting a soluble SOAR domain in a regulated manner, making it a suitable reporter for the STIM1 active state without the downstream cytoplasmic segments including the SOAR domain. ER Ca2+ depletion led to a remarkable release of the SOAR domain from the STIM1 CC1 segment that was reversible after store refilling, which allowed us to investigate the very early events of STIM1 activation preceding the clustering step caused by the liberated SOAR domain. Analysis of the minimal structural features of the CC1–SOAR interaction showed that the very initial segment of the CC1 domain [designated as the α1 segment (Fahrner et al., 2014)] is the main point of contact that is necessary and sufficient to bind the SOAR domain in a controlled manner. This conclusion was in perfect agreement with other recent studies that analyzed the structural requirements of SOAR–CC1 interactions in situ inside living cells using an alternative approach (Fahrner et al., 2014) or by using FRET analysis (Ma et al., 2015). It is important to mention that the ‘4EA’ mutation within the acidic stretch (between 318–322) did not release the SOAR domain from the CC1 clamp in our experiments, and in fact the CC1-α1 segment (STIM1 ending at 280) was sufficient for the regulated capture of the SOAR domain. However, in the context of both the whole STIM1 molecule and the soluble cytoplasmic segment, this mutation clearly weakens the SOAR–CC1 interaction (Korzeniowski et al., 2010; Muik et al., 2011; Zhou et al., 2013). The structure of the entire cytoplasmic STIM1 segment will clarify the mechanism of how these mutations contribute to the destabilization of the intramolecular interaction.
By using this method, we wanted to address the question of whether dimerization or oligomerization is the trigger to break the CC1–SOAR interaction. We found several lines of evidence showing that STIM1 is a dimer even in the resting state. First, cross-linking experiments clearly showed that even STIM1 molecules lacking the SOAR domain in the quiescent state were dimers. Second, FRET analysis suggested that the SOAR domain of one STIM1 molecule can interact with the CC1 domain of another. Third, a forced dimerization of the STIM1–CC1 construct using a BiF approach supported the idea of controlled capture and release of the SOAR domain. To test the effect of oligomerization, we used a strategy relying upon forcing oligomerization of the STIM1 molecules through expression of FRB multimers in the ER lumen along with various FKBP12-tagged STIM1 constructs. Our results suggested that oligomerization alone was not able to evoke activation of STIM1 molecules as long as they had intact luminal domains, yet these constructs responded properly to ER Ca2+ depletion. We found, however, that oligomerization did evoke activation of STIM1 constructs lacking the luminal domains and that such constructs are already in a partially active state. Our results suggested that oligomerization can evoke full activation of STIM1 as long as it contains the SOAR domain and lacks the luminal domains, as observed previously using a different oligomerization approach (Luik et al., 2008). These data suggested that the luminal segments have a major role in keeping the STIM1 molecules in an inactive state and only when this break is released will oligomerization become an important driving force towards full activation.
Based on our results, we propose a model in which STIM1 molecules form constitutive dimers, and their activation bears some analogy to the activation mechanism of growth factor receptors. We suggest that the conformational change in the luminal domains upon Ca2+ unbinding translates to a rotational movement of the two membrane-spanning segments that is transmitted to the CC1 domain. This rotational movement would be sufficient to break the helix–helix interaction that is formed between the STIM1 CC1 and SOAR domains. Our model is fully compatible with the findings and conclusions of the Ma study in that the reorientation of the TM segments is a key step in the early activation cascade. A more recent study from the Zhou laboratory also reached similar conclusions (Ma et al., 2017). Jing et al. has identified an additional player in the control of the CC1 domain SOAR interaction. They discovered that an ER-resident multipass transmembrane molecule they named STIMATE (also known as TMEM110) can also interact with the CC1 domain to weaken its interaction with SOAR (Jing et al., 2015). Although STIMATE appears to be an important modifier of the STIM1 activation process, it probably does not significantly change the conclusions of our study and those of Ma et al. (Ma et al., 2015, 2017).
In summary, we investigated the very initial steps in the STIM1 activation cascade by obtaining dynamic measurements of CC1–SOAR interactions in intact cells combined with forced oligomerization. Based on our findings, we suggest that STIM1 molecules work as dimers, and that the initial step that releases the SOAR domain from an intramolecular clamp upon Ca2+ unbinding is a conformational change in the luminal domains that is transmitted to the initial segment of the cytoplasmic CC1 domain via a rearrangement within the TM segment, perhaps via a rotational movement. Future studies will test the validity of this model.
MATERIALS AND METHODS
Materials
Rapamycin and thapsigargin were obtained from Calbiochem. Lipofectamine 2000 and Fura-2 AM were from Molecular Probes (Invitrogen). Apyrase and ATP were purchased from Sigma. DNA Phusion HF polymerase, Antarctic Phosphatase, T4 DNA ligase and chemically competent E. coli DH5α cells were obtained from New England Biolabs. Restriction enzymes were obtained from New England Biolabs or Fermentas (Thermo Scientific). Cloned DNA Pfu polymerase and the QuikChange site-directed mutagenesis kit were from Stratagene (Agilent). All other chemicals were of the highest analytical grade.
Cell culture, transient transfection, confocal microscopy and fluorescence measurements
COS-7 cells obtained from ATCC and regularly tested for mycoplasma contamination were used for all experiments. Cells were transiently transfected with the indicated constructs (3×105 cells and a total of 1–2 μg of DNA per 35 mm dish, unless stated otherwise) using Lipofectamine 2000 for 24 h as described previously (Korzeniowski et al., 2010). Confocal microscopy was performed with a Zeiss LSM 510 and Zeiss LSM 710 scanning confocal microscopes using a 63×1.4 NA oil immersion objective. Confocal recordings were performed at room temperature in a modified Krebs–Ringer buffer containing 120 mM NaCl, 4.7 mM KCl, 2 mM CaCl2, 0.7 mM Mg(SO4)2, 10 mM glucose, 10 mM Hepes, pH 7.4. Images were analyzed with the Zeiss Zen software. Wide-field microscopy-based FRET experiments were performed as described previously (Korzeniowski et al., 2010). For fluorimetric FRET measurements in cell suspension, an SLM 8100C steady-state fluorimeter (SLM Instruments, Urbana, IL) was used. For this, cells transfected with the indicated STIM1 plasmid DNAs [3×105 cells on a six-well plate transfected with 1 µg DNA (two constructs, 500 ng each)] were harvested by trypsinization and suspended in the modified Krebs–Ringer buffer 24 h after transfection. 106 cells were stirred in an acrylic cuvette at 37°C in Ca2+-free Krebs–Ringer buffer supplemented with 2 mM Ca2+, and the time course of acceptor over donor fluorescence (mApple/AcGFP with excitation at 485 nm, and emission at 515 nm for donor and 580 nm long pass for acceptor) was monitored in response to Ca2+ store depletion with ATP and Tg addition (50 µM and 200 nM, respectively). (Holowka et al., 2014).
Single-cell Ca2+ recordings
Transiently transfected cells were loaded with 3 µM Fura2/AM in HEPES-buffered M199 medium supplemented with 10% pluronic acid and 200 µM sulfinpyrazone as described previously (Korzeniowski et al., 2010). Ca2+ measurements with Fura2-loaded cells were performed in the modified Krebs–Ringer solution at room temperature using an Olympus IX70 inverted microscope equipped with a Lamda-DG4 illuminator and a MicroMAX-1024BFT digital camera and the appropriate filter sets. The MetaFluor (Molecular Devices) software was used for data acquisition and analysis.
DNA constructs
The primers used to generate the different DNA constructs are listed in Table S1. All C-terminally tagged STIM1 constructs were designed based on pEGFP-N1 or pEAcGFP-N1 backbone. Briefly, STIM1 cDNA ware cloned into XhoI and EcoRI restriction sites in pEAcGFP-N1, following the design described by Barr et al. (2008). The C-terminal AcGFP was then replaced with mApple using AgeI and NotI restriction sites. All N-terminally tagged full-length human STIM1 constructs were based on pEYFP-C1 (Clontech) where STIM1 fragments were cloned between EcoRI and KpnI restriction sites as previously described (Korzeniowski et al., 2010). Site-directed mutagenesis was used to introduce desired mutations and to generate deletion constructs with Phusion high-fidelity DNA polymerase or cloned Pfu high-fidelity DNA polymerase. The initial screening was performed with silent mutation introduced into STIM1 in the PCR reaction with new and unique restriction sites. STIM1 constructs with point mutations D76A, L251S and 4EA (E318A, E319A, E320A, E322A) were also used for further cloning steps to prepare the truncated constructs. Mutagenesis products were re-cloned into original backbone after the designed mutations were confirmed by sequencing, and all constructs were fully sequenced to rule out unwanted mutations.
TK-mRFP-FK-STIM1(203-685) was generated in multiple steps based on a mRFP-FK-STIM1 C-terminal piece (Korzeniowski et al., 2010) using HindIII and KpnI restriction sites for STIM1(203–343) cloning and VspI (AseI) and NheI restriction sites to replace the CMV promoter with the TK promoter. A similar procedure was used to generate mRFP–FK–STIM1(203–343) and mRFP–FK–STIM1(1–685) as well as the YFP versions of those constructs. The untagged Orai1 construct was as described previously (Varnai et al., 2007).
BiF GFP-complementation studies were performed with BiGFP Venus. N-terminal 1–155 and C-terminal 155–end fragments were fused to the C-termini of STIM1 (1-343) to complete complementation. For this, BiGFP Venus fragments were first cloned into a pEYFP-N1 vector backbone (Kerppola, 2006). The restriction sites of AgeI and NotI were used for STIM1 cloning using the STIM1(1–343)–mApple construct.
ER luminal (LER) FRB constructs (LER–CFP–FRB) constructs were designed based on pECFP-C1 backbone from Clontech and custom-designed oligonucleotides synthesized by Blue Heron (Bothell, WA). In this design, the opening reading frame (ORF) starts with the calreticulin signal sequence, followed by multiple cloning sites followed by the KDEL ER-retention sequence (shown in Table S2). First, CFP extended with a helical linker was cloned between the NheI and BssHII sites. This was followed by the insertion of amplified FRB fragment with the helical linker into the EcoRV restriction site. The FRB fragment was amplified flanked with EcoRV and SmaI restriction sites, and SmaI was intentionally lost during ligation. Vector backbone 5′ blunt ends were dephosphorylated with Antarctic Phosphatase. Up to five FRB multimers were generated by adding them at the EcoRV site. All constructs were fully sequenced. PM–CFP–FRB multiples were generated by replacing the calreticulin ER-targeting sequence with that of the Lyn kinase N-terminal peptide (Table S2).
Crosslinking experiments
COS-7 cells were plated in six-well dishes and transfected with 0.5 µg STIM1(1–343) or YFP–STIM1(1–280), and 0.2 µg mRFP–SOAR by using Lipofectamine 2000 transfection reagent. Cells were then kept in low-Ca2+ DMEM (0.2 mM Ca2+) for 24 h. Crosslinking experiments were performed the next day. Cells were harvested with mild trypsinization and transferred to nominally Ca2+-free modified Krebs–Ringer solution supplemented with 100 µM EGTA. Cells were treated with ATP and Tg (50 µM and 200 nM, respectively) or solvent for 5 min, then washed and crosslinked with 100 µM disuccinimidyl suberate (DSS; Pierce) for 20 min at room temperature (in 1 ml volume). To quench amino reactive groups 1 ml of 40 mM Tris-HCl pH 7.5 was added (final concentration 20 mM) for 15 min at room temperature. Cells were then washed twice with low-Ca2+ medium and 300 µl of 2× Laemmli buffer was added. Samples were collected, boiled and separated on Nupage 4-12% Tris-Glycine gels. After transfer, membranes were developed with an STIM1 (N-terminus) primary antibody (Sigma-Algrich, S6072 lot# 104M4822 V, dilution 1:3000) and a secondary antibody suitable for imaging in an infrared Odyssey imager (IRDye 680LT, donkey anti-rabbit-IgG conjugated to IRDye 680LT; 926-68023, lot# C51216-05, dilution 1: 10,000).
Native gels
COS-7 cells were plated in six-well dishes and transfected with 0.5 µg YFP–STIM1(1–343), STIM1(1–343)–BiF–Venus [STIM1(1–343)–Venus(1–55)] and STIM1(1–343)–Venus(155–end) or STIM1(1–343)–AcGFP using Lipofectamine 2000 transfection reagent, as described above. After 24 h, cells were washed twice with low-Ca2+ medium and collected in 300 µl of 2× native PAGE loading buffer [40% (v/v) glycerol, 62.5 mM Tris-HCl pH 6.0, 0.01% (w/v) Bromophenol Blue]. Samples were not boiled but pressed through a 26G double-bent needle and separated on Nupage 4–12% Tris-glycine gels. Electrophoresis gels were analyzed in a Storm 860 Phosphorimager (Molecular Dynamics) using the blue fluorescent laser for visualization of the GFP fusion protein band in the gel (excitation at 450 nm and emission light collected with a 520 nm long-pass filter).
Statistical analysis
Calculations ware performed with MS Excel and P values were calculated with a two-tailed and paired t-test distribution function. FRET data points were normalized based on the same-day control experiments with time ‘0’ taken 1 min before activation of store depletion with ATP and Tg treatment. Steady-state values of stimulated FRET responses were taken 5 min after initiation of store depletion. Positive changes in FRET reflect increases in the acceptor:donor fluorescence ratio and negative changes in FRET reflect decreases in this ratio.
Acknowledgements
Confocal imaging was performed at the Microscopy & Imaging Core of the National Institute of Child Health and Human Development, NIH with the kind assistance of Dr Vincent Schram.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.K.K., B.B., D.A.H., T.B.; Methodology: M.K.K., E.W., T.B.; Formal analysis: M.K.K.; Investigation: M.K.K., E.W., T.B.; Writing - original draft: T.B.; Writing - review & editing: M.K.K., B.B., D.A.H., T.B.; Supervision: T.B.; Funding acquisition: B.B., T.B.
Funding
This research was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (NIH) (to M.K.K. and T.B.), and by the National Institute of Allergy and Infectious Diseases (NIH) (R01 AI022449 to M.K.K., B.B. and D.A.H.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.205583.supplemental
References
- Barr V. A., Bernot K. M., Srikanth S., Gwack Y., Balagopalan L., Regan C. K., Helman D. J., Sommers C. L., Oh-Hora M., Rao A. et al. (2008). Dynamic movement of the calcium sensor STIM1 and the calcium channel orai1 in activated T-cells: puncta and distal caps. Mol. Biol. Cell 19, 2802-2817. 10.1091/mbc.E08-02-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloway N., Holowka D. and Baird B. (2010). A basic sequence in STIM1 promotes Ca2+ influx by interacting with the C-terminal acidic coiled coil of Orai1. Biochemistry 49, 1067-1071. 10.1021/bi901936q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington E. D., Wu M. M. and Lewis R. S. (2010). Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol. Biol. Cell 21, 1897-1907. 10.1091/mbc.E10-02-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B., Yang X., Li S., Lin Z., Wang Z., Dong C. and Shen Y. (2013). The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1. PLoS ONE 8, e74735 10.1371/journal.pone.0074735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Wang Y., Zhou Y., Soboloff J. and Gill D. L. (2009). STIM and Orai: dynamic intermembrane coupling to control cellular calcium signals. J. Biol. Chem. 284, 22501-22505. 10.1074/jbc.R109.018655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derler I., Madl J., Schütz G. and Romanin C. (2012). Structure, regulation and biophysics of I(CRAC), STIM/Orai1. Adv. Exp. Med. Biol. 740, 383-410. 10.1007/978-94-007-2888-2_16 [DOI] [PubMed] [Google Scholar]
- Fahrner M., Muik M., Schindl R., Butorac C., Stathopulos P., Zheng L., Jardin I., Ikura M. and Romanin C. (2014). A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1). J. Biol. Chem. 289, 33231-33244. 10.1074/jbc.M114.610022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.-H., Tanasa B., Hogan P. G., Lewis R. S., Daly M. and Rao A. (2006). A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441, 179-185. 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Hogan P. G. and Rao A. (2015). Store-operated calcium entry: Mechanisms and modulation. Biochem. Biophys. Res. Commun. 460, 40-49. 10.1016/j.bbrc.2015.02.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan P. G., Lewis R. S. and Rao A. (2010). Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu. Rev. Immunol. 28, 491-533. 10.1146/annurev.immunol.021908.132550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowka D., Korzeniowski M. K., Bryant K. L. and Baird B. (2014). Polyunsaturated fatty acids inhibit stimulated coupling between the ER Ca(2+) sensor STIM1 and the Ca(2+) channel protein Orai1 in a process that correlates with inhibition of stimulated STIM1 oligomerization. Biochim. Biophys. Acta 1841, 1210-1216. 10.1016/j.bbalip.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. N., Zeng W., Kim J. Y., Yuan J. P., Han L., Muallem S. and Worley P. F. (2006). STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat. Cell Biol. 8, 1003-1010. 10.1038/ncb1454 [DOI] [PubMed] [Google Scholar]
- Jing J., He L., Sun A., Quintana A., Ding Y., Ma G., Tan P., Liang X., Zheng X., Chen L. et al. (2015). Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2+) influx. Nat. Cell Biol. 17, 1339-1347. 10.1038/ncb3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K. (2006). Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278-1286. 10.1038/nprot.2006.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465-487. 10.1146/annurev.biophys.37.032807.125842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski M. K., Manjarres I. M., Varnai P. and Balla T. (2010). Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci. Signal. 3, ra82 10.1126/scisignal.2001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski M. K., Baird B. and Holowka D. (2016). STIM1 activation is regulated by a 14 amino acid sequence adjacent to the CRAC activation domain. AIMS Biophys. 3, 99-118. 10.3934/biophy.2016.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E. Jr. and Meyer T. (2005). STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235-1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Fivaz M., Inoue T. and Meyer T. (2007). Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. USA 104, 9301-9306. 10.1073/pnas.0702866104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik R. M., Wang B., Prakriya M., Wu M. M. and Lewis R. S. (2008). Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature 454, 538-542. 10.1038/nature07065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Wei M., He L., Liu C., Wu B., Zhang S. L., Jing J., Liang X., Senes A., Tan P. et al. (2015). Inside-out Ca(2+) signalling prompted by STIM1 conformational switch. Nat. Commun. 6, 7826 10.1038/ncomms8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Zheng S., Ke Y., Zhou L., He L., Huang Y., Wang Y. and Zhou Y. (2017). Molecular determinants for STIM1 activation during store-operated Ca2+ entry. Curr. Mol. Med. [Epub ahead of print] 10.2174/1566524017666170220103731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M., Fahrner M., Schindl R., Stathopulos P., Frischauf I., Derler I., Plenk P., Lackner B., Groschner K., Ikura M. et al. (2011). STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 30, 1678-1689. 10.1038/emboj.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh A. B. and Putney J. W. Jr. (2005). Store-operated calcium channels. Phys.Rev. 85, 757-810. 10.1152/physrev.00057.2003 [DOI] [PubMed] [Google Scholar]
- Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E. and Lewis R. S. (2009). STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136, 876-890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney J. W., Jr. (1986). A model for receptor-regulated calcium entry. Cell Calcium 7, 1-12. 10.1016/0143-4160(86)90026-6 [DOI] [PubMed] [Google Scholar]
- Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D. et al. (2005). STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435-445. 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J., Rothberg B. S., Madesh M. and Gill D. L. (2012). STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13, 549-565. 10.1038/nrm3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos P. B., Schindl R., Fahrner M., Zheng L., Gasmi-Seabrook G. M., Muik M., Romanin C. and Ikura M. (2013). STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat. Commun. 4, 2963 10.1038/ncomms3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Tóth B., Tóth D. J., Hunyady L. and Balla T. (2007). Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J. Biol. Chem. 282, 29678-29690. 10.1074/jbc.M704339200 [DOI] [PubMed] [Google Scholar]
- Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R. et al. (2006). CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312, 1220-1223. 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Jin H., Cai X., Li S. and Shen Y. (2012). Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA 109, 5657-5662. 10.1073/pnas.1118947109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. P., Zeng W., Dorwart M. R., Choi Y.-J., Worley P. F. and Muallem S. (2009). SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11, 337-343. 10.1038/ncb1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Srinivasan P., Razavi S., Seymour S., Meraner P., Gudlur A., Stathopulos P. B., Ikura M., Rao A. and Hogan P. G. (2013). Initial activation of STIM1, the regulator of store-operated calcium entry. Nat. Struct. Mol. Biol. 20, 973-981. 10.1038/nsmb.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Cai X., Nwokonko R. M., Loktionova N. A., Wang Y. and Gill D. L. (2017). The STIM-Orai coupling interface and gating of the Orai1 channel. Cell Calcium 63, 8-13. 10.1016/j.ceca.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]