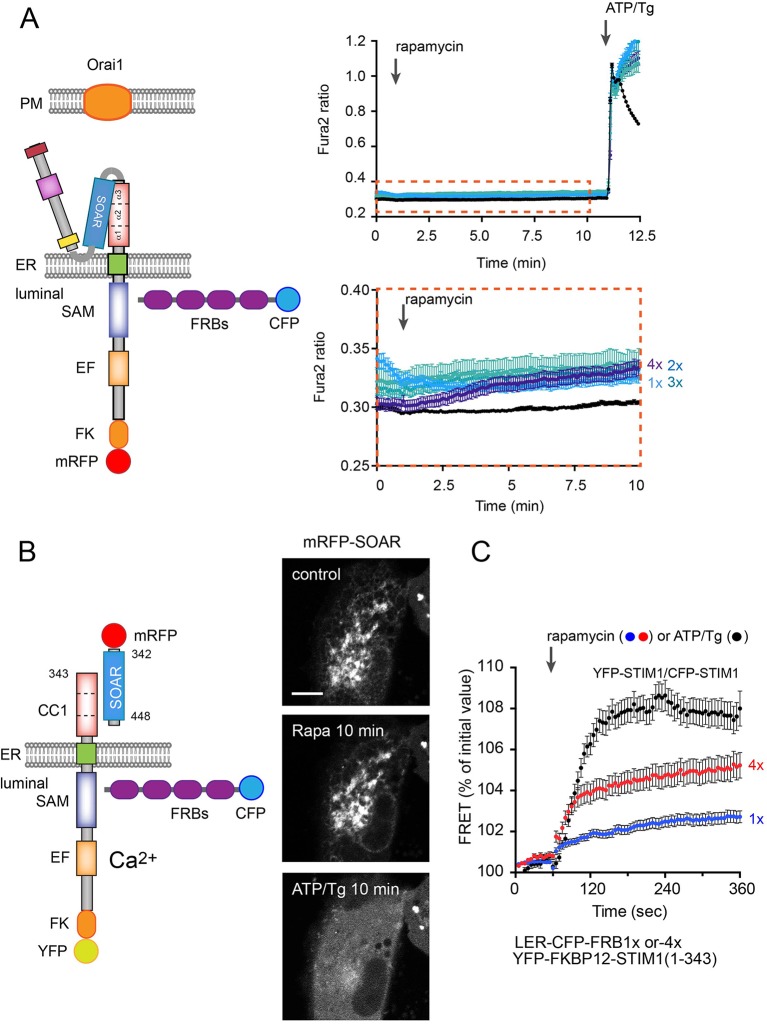
Fig. 6.
The effect of oligomerization on STIM1 activation. Artificial oligomerization was achieved by expressing ER-lumen-targeted FRB multimers (1× to 4×FRB) together with STIM1 proteins that were also tagged with FKBP12 at the lumen. (A) Single-cell cytosolic Ca2+ recordings were performed in transfected cells loaded with Fura2-AM. Cells were transfected with full-length TK-promoter-driven mRFP–FKBP12–STIM1 (wild type), untagged Orai1 and luminally targeted CFP–FRBs. Rapamycin-induced oligomerization induced a minor elevation of cytosolic Ca2+ only when the 4×FRB was present in the ER (see the bottom trace in A with the enlarged scale). This response was very small compared to the one evoked by subsequent store depletion (ATP/Tg treatment). Results are means±s.e.m. from 133–209 cells recorded in 4–7 separate experiments. (B) Rapamycin-driven oligomerization has no detectable effect on the release of the SOAR domain from a YFP–FKBP12–STIM1(1–343) construct, which is then effectively released by ATP/Tg treatment. Scale bar: 10 μm. (C) FRET measurements between the ER-lumen-targeted FRB multimers and YFP–STIM1. Note that the 4×FRBs cause oligomerization that is significantly higher than those evoked by the 1×FRB and ∼50% of the FRET increase shown by STIM1 molecules upon ATP/Tg treatment. Results are means±s.e.m. from 128, 75 and 43 cells for 1×FRB, 4×FRB and the STIM1–STIM1 groups, respectively, obtained in 6–10 independent experiments.