ABSTRACT
The integrity of the cellular proteome is supported by quality control networks, which govern protein synthesis, folding, and degradation. It is generally accepted that an age-related decline in protein homeostasis (proteostasis) contributes to protein aggregation diseases. However, the mechanistic principles underlying proteostasis imbalance and the impact on life expectancy are not well understood. We recently demonstrated that this interrelation is affected by chaperone-directed ubiquitylation, shifting the amount of the conserved DAF-2/insulin receptor both in Caenorhabditis elegans and Drosophila melanogaster. The ubiquitin ligase CHIP either targets the membrane bound insulin receptor or misfolded proteins for degradation, which depends on the cellular proteostasis status. Increased proteotoxicity triggers chaperone-assisted redirection of CHIP toward protein aggregates, limiting its capacity to degrade the insulin receptor and prevent premature aging. In light of these findings, we discuss a new concept for understanding the impact of proteome imbalance on longevity risk.
KEYWORDS: aging, C. elegans, CHIP, CHN-1, DAF-2, insulin signaling, molecular chaperone, proteostasis, ubiquitin
Proteostasis and aging are intricately balanced
The proteome is defined as the entire set of proteins expressed in a given cell-type or organism, which can vary with time and physiologic status.1 The integrity of the proteome is maintained through numerous quality control pathways, which form a complex proteostasis network. The coordination between the different proteostatic nodes is tightly balanced and adjusted in response to proteotoxic stress caused by environmental and metabolic challenges.2,3 The human proteostasis network involves >1000 accessory factors and regulatory components, which govern protein synthesis, folding, and degradation.3 For example, molecular chaperones support efficient folding of nascent polypeptides synthesized at the ribosome to secure their biologic function(s).1 Otherwise, defective folding could result in increased abundance of toxic protein aggregates, which endanger the integrity of the entire proteome.1 Molecular chaperones additionally participate in refolding of damaged proteins that accumulate upon proteotoxic stress conditions. In case protein refolding cannot be sufficiently executed, chaperones team up with the ubiquitin/proteasome-system (UPS) or autophagy pathway to trigger degradation of misfolded proteins.4-8 The UPS is one major proteolytic component of the cellular proteostasis network mediating the degradation of regulatory or damaged proteins.8,9 Turnover by the 26S proteasome is highly selective and initiated by covalent attachment of the small, evolutionarily conserved protein ubiquitin predominantly to internal lysine residues.10 Recent studies in different organisms supported the idea that the activity of the 26S proteasome progressively declines during aging, although molecular aspects of this regulation have not been addressed.11 One limiting factor important for proteasomal activity in worms and human embryonic stem cells is the 19S regulatory particle (RP) subunit RPN-6/PSMD11, as its overexpression causes increased proteasome assembly, proteotoxic stress resistance, and lifespan extension.12
The most extensively studied genetic program regulating stress tolerance and longevity is the insulin/insulin-like growth factor 1 (IGF-1) signaling (IIS).13 The central regulator of the conserved IIS pathway in C. elegans is the insulin receptor DAF-2, which is important for lifespan, stress responses, and metabolism.13 Loss of DAF-2 signaling induces nuclear translocation and activation of the downstream FOXO transcription factor DAF-16, which regulates gene expression to enhance stress resistance and lifespan14,15 including molecular chaperones and proteasomal subunits like RPN-6 (Fig. 1).12,16-18 The current literature on IIS indicates that proteostasis and aging are inextricably linked to stress tolerance. Surprisingly, however, recent proteomic approaches performed in C. elegans did not show any age-related activity decline of the protein degradation machinery.19 Whereas protein synthesis decreased, protein degradation pathways seem to be more efficient. This result contradicts the general concept of proteostasis collapse explained by reduced proteasomal turnover of misfolded proteins. In the course of aging, errors in protein synthesis pathways increase, which is caused by genomic instability,20 an accumulation of transcription errors,21 mistranslation, and misfolding.22,23 These hallmarks of aging lead to the accumulation of misfolded proteins and finally to the formation of protein aggregates. Here, we discuss an alternative concept for understanding the complex interplay between proteostasis and aging, highlighting an active role of misfolded proteins, which ultimately limit the capacity of protein quality control networks and further accelerate the aging process (Fig. 2).
Figure 1.
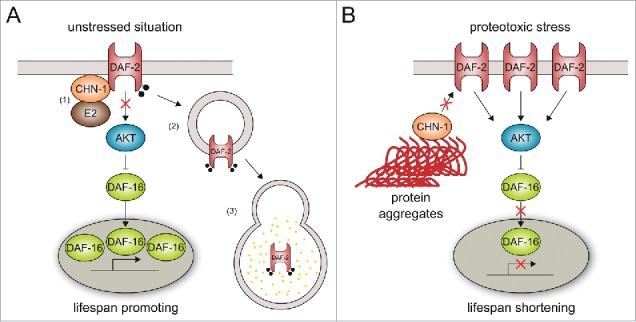
CHN-1 defines insulin signaling and longevity through DAF-2 ubiquitylation. (A) In the absence of stress, (1) CHN-1 binds to and monoubiquitylates the DAF-2/insulin receptor in collaboration with an E2 enzyme (e.g. LET-70),24 which triggers (2)-(3) endocytic-lysosomal degradation of the insulin receptor.6 Reduced insulin signaling supports nuclear localization of the transcription factor DAF-16 and expression of pro-longevity genes. (B) Stress- or aging-related decline in proteostasis cause increased level of misfolded proteins, which redirects CHN-1 activity toward chaperone-assisted ubiquitylation. The stabilization of membrane-bound DAF-2 triggers insulin signaling, which limits nuclear translocation of DAF-16 and consequently shortens lifespan.
Figure 2.
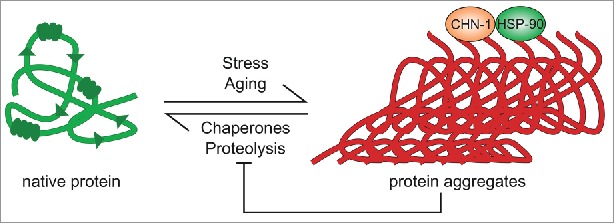
Proteostasis collapse redirects substrate specificities of the proteolytic network. Stress and aging induce a rearrangement of protein quality control networks, including molecular chaperones and protein degradation pathways, to maintain proteostasis. Among other quality control factors, this process recruits the E3 ubiquitin ligase CHN-1 and co-working chaperone Hsp90 to protein aggregates for chaperone-assisted ubiquitylation of misfolded proteins. Consequently, the activity of natural CHN-1 substrates remains uncontrolled, which provokes pathologies and further accelerates aging.
The quality control ubiquitin ligase CHIP – A central regulator of proteostasis
The C terminus of Hsc70-interacting protein (CHIP) was originally identified as a binding partner of molecular chaperones.24 The direct interaction between the E3 ligase CHIP and Hsp70 or Hsp90 facilitates chaperone-assisted ubiquitylation of misfolded proteins that are bound to the chaperone complex. This intricate cooperation between both chaperone and proteasome systems facilitates the cellular balance of protein folding and degradation and maintains the cellular proteostasis network.25-27 Besides proteasomal turnover, CHIP-dependent ubiquitylation of damaged proteins also triggers disposal through endocytic-lysosomal pathways,28,29 and autophagy.30 In agreement with its central role in proteostasis, CHIP prevents an age-related pathological accumulation of protein aggregates.25,31 CHIP directly modulates the proteotoxic stress response by activating heat shock factor 1 (HSF-1) and by reducing the level of Hsp70 after heat shock.32,33 However, the molecular mechanism that determines between refolding or destruction of chaperone substrates is not fully understood.
CHIP is conserved and orthologs exist in worms, flies, fish, mammals, and even plants. The worm ortholog CHN-1 has been shown to team up with another E3 ligase called UFD-2 to target the myosin chaperone UNC-45 for degradation.24 Here, the E3/E4 ligase complex regulates the assembly and maintenance of myosin filaments in striated C. elegans muscle. In agreement with its role in protein quality control, CHIP knockout mice exhibit reduced lifespan associated with age-related pathophysiological defects.34 Otherwise, CHIP knockout mice exhibit normal embryonic development and unaffected turnover of various CHIP substrates.34,35 These physiologic characteristics indicate functional redundancy among different quality control E3 ligases and the existence of at least one critical CHIP-specific substrate that restricts longevity. Due to the fact that CHIP exists in a variety of eukaryotic organisms, its role in aging regulation may be evolutionarily conserved. Indeed, CHIP triggers degradation of the insulin receptor (INSR), which regulates metabolic changes and determines lifespan in worms, flies, and human cells.6
CHIP regulates the turnover of the insulin receptor
IIS is well known to affect both lifespan and metabolism in C. elegans.36 We have recently shown that the ubiquitin ligase CHIP either mediates endocytic-lysosomal degradation of the INSR or collaborates with chaperones to degrade misfolded and aggregated proteins.6 Deletion of the C. elegans homolog chn-1 shortens the lifespan and reduces the body size of worms. Although these deletion phenotypes point toward limited nutrient uptake, CHN-1 acts independently of dietary restriction.37 chn-1 mutants show a delayed nuclear transport of DAF-16 leading to reduced expression of longevity genes, which indicates regulation of DAF-2/INSR signaling (Fig. 1). Moreover, depletion of Drosophila CHIP (dCHIP) causes increased phosphorylation of AKT kinase,38 which is another indication for activated insulin signaling. CHIP directly interacts with and monoubiquitylates multiple lysine residues of the INSR with Lys1047 being the main ubiquitylation site, also identified in an unbiased proteomic study.39 Substitution of Lys1047 (Lys1047Arg) elevates the INSR level, indicating that CHIP-dependent ubiquitylation at this site is crucial for endocytic-lysosomal degradation of the receptor. Indeed, down regulation of chn-1, dCHIP, and human STUB1 leads to a stabilization of DAF-2/INSR, both in C. elegans, Drosophila, and HEK293T cells, respectively. In contrast to INSR regulation, CHIP does not ubiquitylate the insulin-like growth factor receptor 1, which underlines its substrate specificity.
According to its role as quality control E3 ligase, overexpression of aggregation-prone polyglutamine (polyQ) proteins fosters recruitment of CHIP toward inclusion bodies, thereby reducing the free pool of CHIP. Consequently, CHIP activity is shifted from INSR binding to chaperone-assisted ubiquitylation of misfolded proteins, causing stabilization of the membrane-bound INSR, increased insulin signaling, and reduced expression of longevity genes (Fig. 1). To confirm the tight interplay between CHIP-mediated proteostasis and lifespan regulation we tested the impact of proteotoxic stress on proteome stability and longevity. In fact, chn-1 deletion worms are highly sensitive to paraquat treatment especially during late stages of adulthood. In line with this observation, depletion of dCHIP leads to a dramatic accumulation of oxidatively damaged proteins in aged flies. Oxidative stress causes stabilization of the insulin receptor, which is most likely a result of limited ubiquitin ligase activity because degradation of the INSR can be restored by CHIP overexpression. Given its role in chaperone-assisted ubiquitylation, it is intriguing to note that CHN-1 is mainly required during aging to confer heat stress resistance in worms. Our observation might be linked to an age-related decline in proteostasis known to cause high abundance of misfolded proteins,40-42 which would redirect CHN-1 activity toward the degradation of misfolded proteins, especially during aging. In fact, worms shifted to high temperature at the beginning of adulthood show elevated levels of DAF-2 protein similar to chn-1 lacking mutants grown at normal conditions. Moreover, the shifted worms exhibit shortened lifespan, reflecting increased insulin signaling based on DAF-2 stabilization.
These intriguing findings shed new light on the mechanism by which proteostasis dysfunction accelerates the aging process, showing that there is a significant change in substrate preferences of key regulatory proteolytic pathways, directed toward toxic protein aggregates (Fig. 2). As a consequence, metabolic signaling pathways like IIS are “let off the leash” resulting in physiologic changes and lifespan reduction.
Future directions – mechanisms modifying CHIP activity and specificity
The role of CHIP in chaperone-directed ubiquitylation of non-native proteins is well known to prevent an age-dependent pathological accumulation of protein folding stress. Our work identified a novel function of CHIP in lifespan control, which defines INSR abundance and consequently activity of IIS.6 Since CHIP level remain unchanged even upon proteotoxic stress conditions, CHIP activity is bypassed toward toxic protein aggregates during aging (Fig. 2). This competitive rewiring of quality control pathways results in severe metabolic changes linked to increased IIS, accelerating proteostasis collapse and the aging process (Fig. 1B). Both spatiotemporal control of CHIP function as well as substrate specificity and processing remain to be further addressed. It is tempting to speculate that ligase activity and substrate binding might be coordinated by post-translational modifications of CHIP.43 Interestingly, CHIP is differently ubiquitylated by the E2 enzymes UbcH5A or Ube2W.44,45 Otherwise the neuroprotective function of CHIP is negatively affected by Cdk5-dependent phosphorylation.46
At physiologic concentrations, CHIP proteins from different species form homodimers.47 The structure of dimeric mouse CHIP reveals an unusual asymmetry in which the protomers adopt radically different conformations48 and breaking of symmetry during homodimeric assembly induces E3 ubiquitin ligase activity.49 As a consequence of this asymmetric arrangement, one of the 2 catalytical U-box domains is sterically hindered and not accessible for E2 enzyme binding. Thus, the asymmetric structure of the CHIP dimer provides an elegant means for coupling a dimeric chaperone to a single ubiquitylation system, supporting the formation of a monotonic polyubiquitin chain. In line with the suggested structural model for CHIP mediated ubiquitylation, dimerization of CHIP might determine E2 enzyme binding, ubiquitin chain topology, and substrate selection. Indeed, a first attempt has been made to test how disease-related mutations affect the molecular architecture and the activity of CHIP.50 Deciphering the molecular mechanisms underlying the regulation of CHIP activity and substrate specificity might contribute to develop novel therapeutic strategies directed against age-related diseases including diabetes, cancer, and neurodegenerative disorders.
Abbreviations
- CHIP
C terminus of Hsc70-interacting protein
- HSF-1
heat shock factor 1
- HSP
heat-shock protein
- INSR
insulin receptor
- IIS
insulin/IGF-1 signaling
- IGF-1
insulin-like growth factor 1
- polyQ
polyglutamine
- PTM
post-translational modification
- RP
regulatory particle
- SCAR16
autosomal recessive spinocerebellar ataxia
- TPR
tetratricopeptide repeat
- UPS
ubiquitin/proteasome-system
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of our laboratory for crucial discussion and helpful advice on the manuscript. We apologize for not having cited valuable contributions due to size limitation.
Funding
This work is supported by the European Research Council (consolidator grant 616499) to T.H. In addition, this article is based upon work from COST Action (PROTEOSTASIS BM1307).
References
- [1].Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. PMID:27365453 [DOI] [PubMed] [Google Scholar]
- [2].Noormohammadi A, Calculli G, Gutierrez-Garcia R, Khodakarami A, Koyuncu S, Vilchez D. Mechanisms of protein homeostasis (proteostasis) maintain stem cell identity in mammalian pluripotent stem cells. Cell Mol Life Sci. 2017;doi: 10.1007/s00018-017-2602-1. PMID:28748323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959-91. doi: 10.1146/annurev.biochem.052308.114844. PMID:19298183. [DOI] [PubMed] [Google Scholar]
- [4].Kettern N, Dreiseidler M, Tawo R, Höhfeld J. Chaperone-assisted degradation: Multiple paths to destruction. Biol Chem. 2010;391:481-9. doi: 10.1515/bc.2010.058. PMID:20302520 [DOI] [PubMed] [Google Scholar]
- [5].Kevei É, Pokrzywa W, Hoppe T. Repair or destruction-an intimate liaison between ubiquitin ligases and molecular chaperones in proteostasis. FEBS Lett. 2017;1-20; PMID:28699655; https://doi.org/ 10.1002/1873-3468.12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tawo R, Pokrzywa W, Kevei E, Akyuz ME, Balaji V, Adrian S, Höhfeld J, Hoppe T. The Ubiquitin Ligase CHIP Integrates Proteostasis and Aging by Regulation of Insulin Receptor Turnover. Cell. 2017;169:470-82. doi: 10.1016/j.cell.2017.04.003. PMID:28431247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ulbricht A, Höhfeld J. Tension-induced autophagy: May the chaperone be with you. Autophagy. 2013;9:920-2. doi: 10.4161/auto.24213. PMID:23518596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vilchez D, Saez I, Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun. 2014;5:5659. doi: 10.1038/ncomms6659. PMID:25482515 [DOI] [PubMed] [Google Scholar]
- [9].Dikic I. Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem. 2017;86:1-32. doi: 10.1146/annurev-biochem-061516-044908. PMID:28125288 [DOI] [PubMed] [Google Scholar]
- [10].Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic Mechanism and Assembly of the Proteasome Catalytic Mechanism and Assembly of the Proteasome. Chem Rev. 2009;109:1509-36. doi: 10.1021/cr8004857. PMID:19265443 [DOI] [PubMed] [Google Scholar]
- [11].Hoppe T. Life and destruction: ubiquitin-mediated proteolysis in aging and longevity. F1000 Biol Rep 2010; 2:79. doi: 10.3410/B2-79. PMID:21151840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues APC, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263-8. doi: 10.1038/nature11315. PMID:22922647 [DOI] [PubMed] [Google Scholar]
- [13].Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an Insulin Receptor – Like Gene That Regulates Longevity and Diapause in Caenorhabditis elegans. Science 277(5328):942-946; PMID:9252323; https://doi.org/ 10.1126/science.277.5328.942 [DOI] [PubMed] [Google Scholar]
- [14].Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. Elegans. Nature. 1997;389:994-9. doi: 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- [15].Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139-45. doi: 10.1038/88850. PMID:11381260 [DOI] [PubMed] [Google Scholar]
- [16].Murphy CT. The search for DAF-16/FOXO transcriptional targets: Approaches and discoveries. Exp Gerontol. 2006;41:910-21. doi: 10.1016/j.exger.2006.06.040. PMID:16934425 [DOI] [PubMed] [Google Scholar]
- [17].Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479-87. doi: 10.1242/jcs.001222. PMID:17646672. [DOI] [PubMed] [Google Scholar]
- [18].Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct Inhibition of the Longevity-Promoting Factor SKN-1 by Insulin-like Signaling in C. elegans. Cell 2008;132:1025-38. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto RI, Dobson CM, Vendruscolo M, Mann M, et al.. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell. 2015;161:919-32. doi: 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194-1217; PMID:23746838; doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vermulst M, Denney AS, Lang MJ, Hung C-W, Moore S, Mosely AM, Thompson WJ, Madden V, Gauer J, Wolfe KJ, et al.. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat Commun. 2015;6:8065. doi: 10.1038/ncomms9065. PMID:26304740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715-24. doi: 10.1038/nrg2662. PMID:19763154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ling J, Cho C, Guo LT, Aerni HR, Rinehart J, Söll D. Protein Aggregation Caused by Aminoglycoside Action Is Prevented by a Hydrogen Peroxide Scavenger. Mol Cell. 2012;48:713-22. doi: 10.1016/j.molcel.2012.10.001. PMID:23122414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hoppe T, Cassata G, Barral JM, Springer W, Hutagalung AH, Epstein HF, Baumeister R. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118:337-49. doi: 10.1016/j.cell.2004.07.014 [DOI] [PubMed] [Google Scholar]
- [25].Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93-6. doi: 10.1038/35050618. PMID:11146632 [DOI] [PubMed] [Google Scholar]
- [26].Cyr DM, Höhfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368-75. doi: 10.1016/S0968-0004(02)02125-4. PMID:12114026 [DOI] [PubMed] [Google Scholar]
- [27].Murata S, Chiba T, Tanaka K. CHIP: A quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol. 2003;35:572-8. doi: 10.1016/S1357-2725(02)00394-1. PMID:12672450 [DOI] [PubMed] [Google Scholar]
- [28].Okiyoneda T, Barrière H, Bagdány M, Rabeh WM, Du K, Höhfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805-10. doi: 10.1126/science.1191542. PMID:20595578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Slotman JA, Da Silva Almeida AC, Hassink GC, Van De Ven RHA Van Kerkhof P, Kuiken HJ, Strous GJ. Ubc13 and COOH terminus of Hsp70-interacting protein (CHIP) are required for growth hormone receptor endocytosis. J Biol Chem. 2012;287:15533-43. doi: 10.1074/jbc.M111.302521. PMID:22433856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, et al.. Chaperone-Assisted Selective Autophagy Is Essential for Muscle Maintenance. Curr Biol. 2010;20:143-8. doi: 10.1016/j.cub.2009.11.022. PMID:20060297 [DOI] [PubMed] [Google Scholar]
- [31].Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, et al.. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703-14. doi: 10.1093/hmg/ddh083. PMID:14962978 [DOI] [PubMed] [Google Scholar]
- [32].Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, et al.. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446-58. doi: 10.1093/emboj/cdg529. PMID:14532117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Qian S-B, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551-5. doi: 10.1038/nature04600. PMID:16554822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28:4018-25. doi: 10.1128/MCB.00296-08. PMID:18411298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942-52. doi: 10.1093/hmg/ddn296. PMID:18784277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Perez CL, Van Gilst MR. A 13C Isotope Labeling Strategy Reveals the Influence of Insulin Signaling on Lipogenesis in C. elegans. Cell Metab. 2008;8:266-74. doi: 10.1016/j.cmet.2008.08.007 [DOI] [PubMed] [Google Scholar]
- [37].Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091-6. doi: 10.1073/pnas.95.22.13091. PMID:9789046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Su C, Wang C, Lan K, Li C, Chao Y, Lin H, Lee S, Lee W. Akt phosphorylation at Thr308 and Ser473 is required for CHIP-mediated ubiquitination of the kinase. Cell Signal. 2011;23:1824-30. doi: 10.1016/j.cellsig.2011.06.018. PMID:21767636 [DOI] [PubMed] [Google Scholar]
- [39].Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al.. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325-40. doi: 10.1016/j.molcel.2011.08.025. PMID:21906983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427-38. doi: 10.1101/gad.1657108. PMID:18519635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914-9. doi: 10.1073/pnas.0902882106. PMID:19706382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21:1406-15. doi: 10.1038/nm.4001. PMID:26646497. [DOI] [PubMed] [Google Scholar]
- [43].Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938-44. doi: 10.1074/jbc.M101968200. PMID:11557750 [DOI] [PubMed] [Google Scholar]
- [44].Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723-9. doi: 10.1042/BJ20071338. PMID:18042044 [DOI] [PubMed] [Google Scholar]
- [45].Tatham MH, Plechanovová A, Jaffray EG, Salmen H, Hay RT. Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem J. 2013;453:137-45. doi: 10.1042/BJ20130244. PMID:23560854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kim C, Yun N, Lee J, Youdim MBH, Ju C, Kim W-K, Han P-L, Oh YJ. Phosphorylation of CHIP at Ser20 by Cdk5 promotes tAIF-mediated neuronal death. Cell Death Differ. 2016;23:333-46. doi: 10.1038/cdd.2015.103. PMID:26206088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nikolay R, Wiederkehr T, Rist W, Kramer G, Mayer MP, Bukau B. Dimerization of the Human E3 Ligase CHIP via a Coiled-coil Domain Is Essential for Its Activity. J Biol Chem. 2004;279:2673-8. doi: 10.1074/jbc.M311112200. PMID:14610072 [DOI] [PubMed] [Google Scholar]
- [48].Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation – Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525-38. doi: 10.1016/j.molcel.2005.09.023. PMID:16307917 [DOI] [PubMed] [Google Scholar]
- [49].Ye Z, Needham PG, Estabrooks SK, Whitaker SK, Garcia BL, Misra S, Brodsky JL, Camacho CJ. Symmetry breaking during homodimeric assembly activates an E3 ubiquitin ligase. Sci Rep. 2017;7:1789. doi: 10.1038/s41598-017-01880-4. PMID:28496195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pakdaman Y, Sanchez-Guixé M, Kleppe R, Erdal S, Bustad HJ, Bjørkhaug L, Haugarvoll K, Tzoulis C, Heimdal K, Knappskog PM, et al.. In vitro characterization of six STUB1 variants in spinocerebellar ataxia 16 reveals altered structural properties for the encoded CHIP proteins. Biosci Rep. 2017;37:BSR20170251. doi: 10.1042/BSR20170251. PMID:28396517 [DOI] [PMC free article] [PubMed] [Google Scholar]


