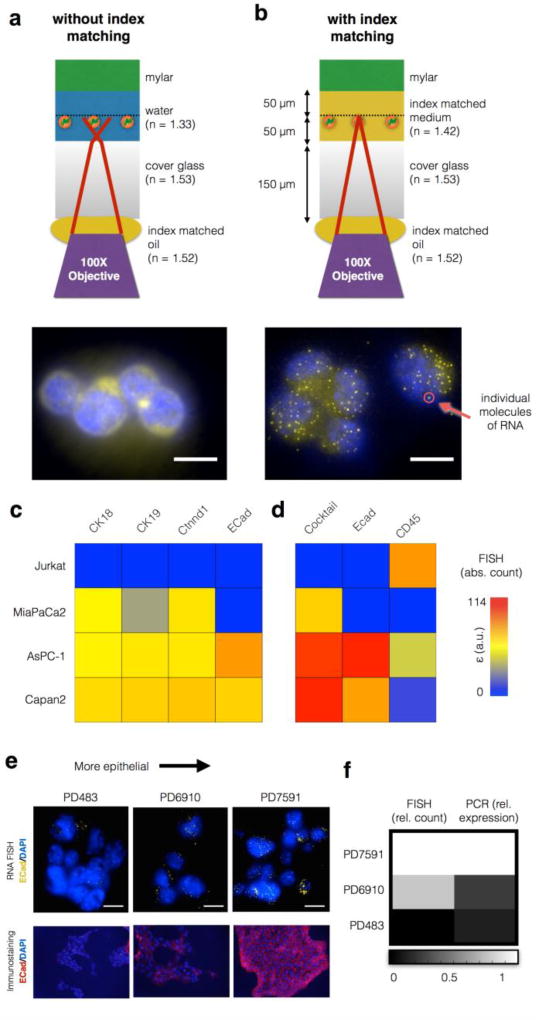
Figure 4. Single Molecule In Situ RNA analysis on the CaTCh FISH chip.
With a 100× objective, individual mll1 RNA molecules were impossible to resolve without index matching (a), but became easily resolved with index matching (b). Scale bar:10 µm. c. RNA FISH on the CaTCh FISH chip was used to enumerate the number of RNA in individual cells for a variety of cell types, represented in a heat map. d. By using a cocktail of RNA markers (CK18, CK19, and Ctnnd1) and CD45 as a negative marker, all of the cell types can be identified. Additionally, ECad can be used to analyze the EMT state of the cell. e. Comparison of RNA FISH and immunofluorescence for ECad in PD483 (mesenchymal), PD6910 (intermediate), and PD7591 (epithelial) cultured cells. Scale bar is 10 µm. f. Comparison of Turbo FISH and quantitative PCR results for the cell lines in (e).