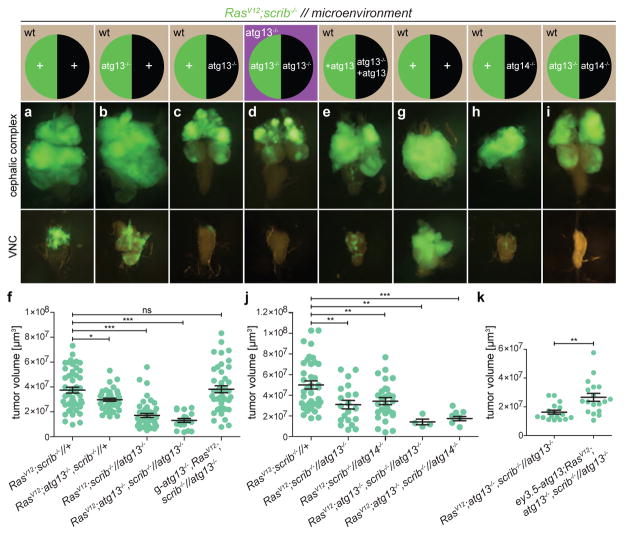
Figure 2. Local NAA is required for tumour growth and invasion.
a–e, g–i, Cartoon (top)illustrates the genotype of EAD (circle, tumour cells in green, microenvironment in black) and other somatic tissues (square). Fluorescent images of tumour growth and invasion (green) in the indicated tissues at day 8 or day 10 is shown below. a depicts RasV12scrib−/− tumours and b depicts tumours with tumour cells deficient in atg13. c depicts RasV12scrib−/− tumours surrounded by an atg13-null microenvironment whereas d depicts an entirely atg13-null animal. e depicts an atg13-null EAD complemented with transgenic atg13, rescuing tumour growth. f, Quantification of tumour volumes at day 8. n = 49 (RasV12scrib−/−//+); n = 42 (RasV12atg13−/−scrib−/−//+); n = 45 (RasV12scrib−/−//atg13−/−); n = 18 (RasV12atg13−/−scrib−/−//atg13−/−); n = 43 (g-atg13+RasV12scrib−/−//atg13−/−). g depicts RasV12scrib−/− tumours at day 10, h shows the same but with atg14-null cells in the microenvironment, whereas i shows atg13-deficient tumours confronted with atg14−/− cells in the microenvironment. j, Quantification of tumour volumes of the indicated genotypes at day 8. n = 38 (RasV12scrib−/−//+). n = 20 (RasV12scrib−/−//atg13−/−). n = 28 (RasV12scrib−/− //atg14−/−). n = 4 (RasV12atg13+scrib−/−//atg13−/−). n = 8 (RasV12atg13−/−scrib−/− //atg14−/−). k, Tumour volumes with or without EAD-specific rescue construct ey3.5-atg13 at day 10. n = 17 (RasV12atg13−/−scrib−/−//atg13−/−). n = 18 (ey3.5-atg13+RasV12atg13−/−scrib−/−//atg13−/−). Values depict mean ± s.e.m. of three independent pooled experiments (f, j, k). NS, not significant, *P ≤ 0.05, **P ≤ 0.01, ***P < 0.0001 from one-way ANOVA with Dunnett’s correction (f, j). **P = 0.0011 from unpaired two-tailed t-test (k).