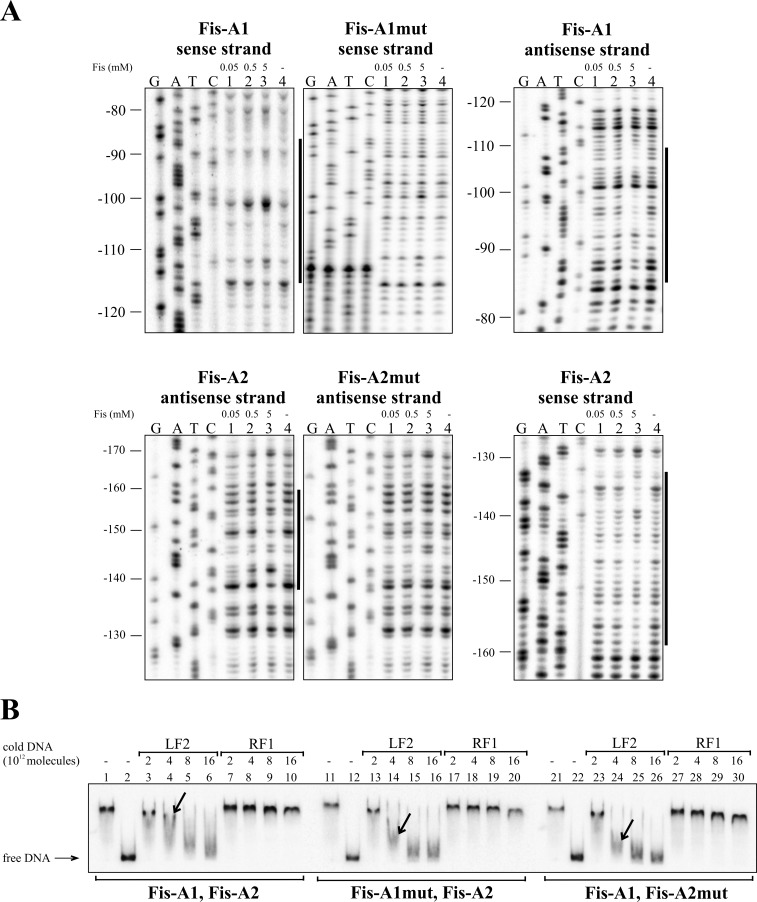
Fig 4. Fis binding to Fis-A1 and Fis-A2 sites upstream of the lapA gene.
(A) Protection of the lapA upstream DNA against DNase I cleavage by Fis binding on the sense and the antisense strands. Lines at the right side of the panels indicate the regions protected by Fis from DNase I cleavage at the positions -110 to -85 on the sense strand and -109 to -84 on the antisense strand corresponding to Fis-A1; and -158 to -132 on the sense strand and -159 to -138 on the antisense strand corresponding to Fis-A2. (B) Gel shift assay of the Fis binding to the lapA promoter DNA containing the wild-type Fis binding site Fis-A1 and Fis-A2 and one or the other mutated site. 2 × 1010 molecules of radioactively labelled PCR products containing both Fis binding sites Fis-A1 and FisA2 or one mutated binding site FisA1mut-Fis-A2, and Fis-A1-FisA2mut were used in Fis binding assay. Fis was outcompeted from Fis-DNA complex with unlabelled PCR product containing the Fis binding site (LF2) or a PCR product without Fis binding site (RF1). Arrows point to different dissociation of Fis from radioactively labelled DNA in favour of binding unlabelled Fis-specific DNA. Added unlabelled DNA was calculated in molecules. 0.46 μM Fis was used in each reaction mixture except mixtures without Fis in lanes 2, 12 and 22.