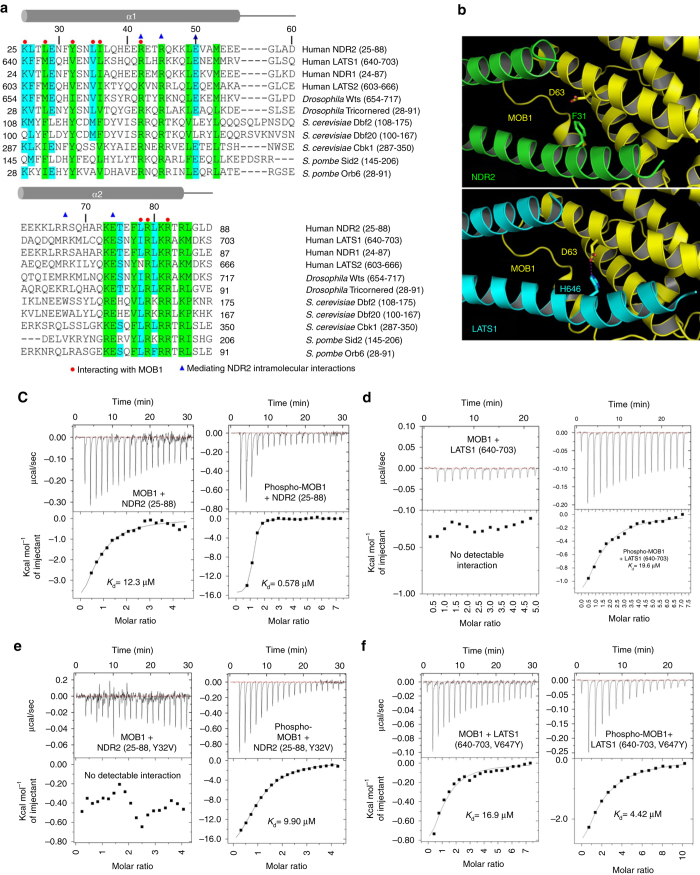
Fig. 2.
MOB1 binds differently to the NTR domains of NDR2 and LATS1 through key residues. a Structure based sequence alignment of the conserved N-terminal regulatory domain of NDR/LATS kinases that is required for MOB1 binding. NDR2 residues mediating interactions with MOB1 are marked with red circles. Residues mediating intramolecular interactions of NDR2 are denoted by blue triangles. Residues conserved from yeast to humans are highlighted in green, while residues conserved in at least seven of the conserved NDR/LATS family members are marked in cyan. α helices are indicated with gray rods. b Asp63 of MOB1 is involved in LATS1 binding, but does not contribute to the interaction of MOB1 with NDR2. MOB1 is shown in yellow, while NDR2 and LATS1 are indicated in green and cyan, respectively. Top panel, Phe31 of NDR2 does not interact with Asp63 of MOB1. Bottom panel, His646 of LATS1 forms a hydrogen bond with Asp63 of MOB1. c–f Isothermal titration calorimetry (ITC) assays measuring the dissociation constant (K d) of indicated non-phosphorylated full-length MOB1 or MST2-phosphorylated MOB1 (phospho-MOB1) with wild-type and mutant NDR2 (25–88) or LATS1 (640–703) variants. MOB1/NDR2 complex formation was dramatically increased by prior phosphorylation of MOB1 c. ITC measurements could not detect any interaction between LATS1 wild-type and non-phosphorylated MOB1, while phospho-MOB1 bound to LATS1 d. The NDR2(Y32V) mutant did not associate with non-phosphorylated MOB1, but bound to phospho-MOB1, although with a 20-fold decreased binding affinity compared to wild-type NDR2 (e). The mutation of Val647 to Tyr of LATS1 enabled LATS1 to bind to non-phosphorylated MOB1, and LATS1(V647Y) displayed a 5-fold increased binding affinity for phospho-MOB1 compared to wild-type LATS1 (f)