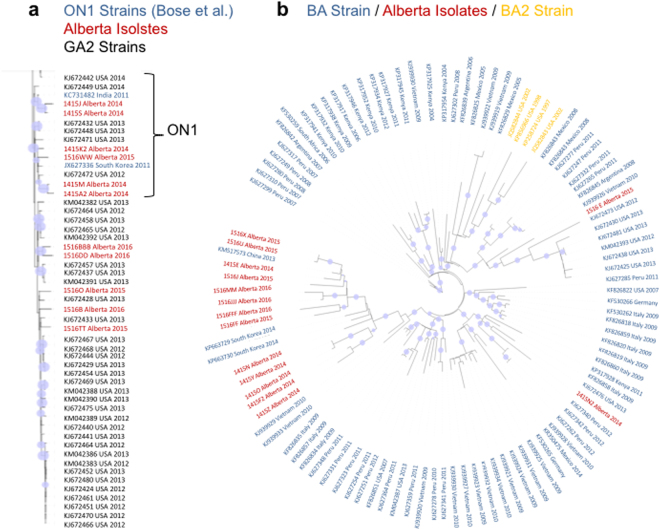
Figure 4.
Comparison of RSV phylogenetics of Alberta, Canada patient NP sample titres during the 2014–2015 and 2015–2016 seasons to RSV strains that have been published in the NCBI database. The phylogenetic tree was created using RAxML software from an original MUSCLE alignment of the complete genomes. Reference strains previously published or identified from known genotypes obtained from the NCBI genome database are indicated above the trees. Tree topology were supported by bootstrap analysis. The blue circles indicate 100% bootstrap confidence. The tree was visualized using a Portable Network Graphics file that was downloaded into Geneious software. (a) A subset of a complete unrooted tree from all RSV type-A genotypes published in NCBI along with Alberta strains obtained for this study. The entire tree was too large to visualize our isolates in the context of the NCBI database, therefore we zoomed into the clade containing Alberta isolates (shown in Red). (b) The unrooted tree from a complete download of all RSV type-B genotypes published in NCBI. Alberta isolates are indicated in red. All isolates were post 2004 therefore the tree only shows the BA strains and the BA2 clade superseding it.