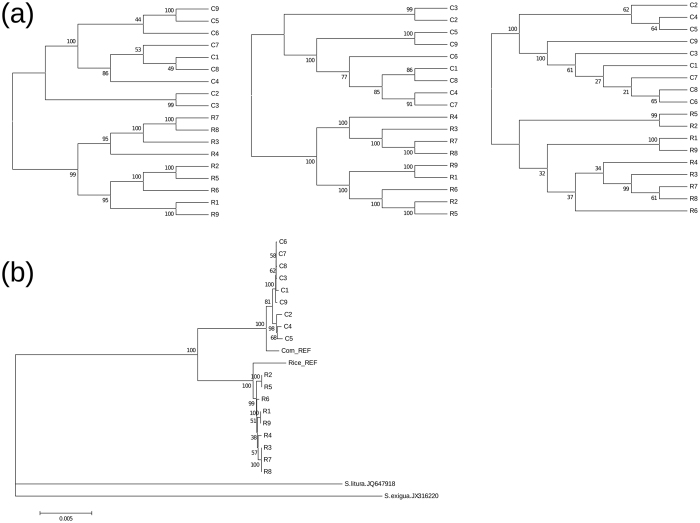
Figure 3.
Phylogenetic relationship among individuals. (a) Neighbour joining phylogenetic trees of the mapping of resequencing data from natural samples of corn strains (from MS_C1 to MS_C8) and rice strain (from MS_R1 to MS_R8) against the reference genomes of the C strain (left), the R strain (middle) and mitochondrial DNA (right). The average genetic distance between pairs of individuals was estimated by the comparison of the genotype (see supplementary information for mode detail) and the distance matrix was generated from these distances. The neighbour joining tree was reconstructed using neighbour program in the phylip package with 1,000 bootstrapping, and the consensus tree was generated using the consense program in the same package. (b) Neighbour joining phylogenetic tree of mitochondrial genomes from natural populations of the corn (C1-C9) and the rice (R1-R9), reference sequences of the corn (Corn REF) and the rice strains (Rice REF) and outgroup species (Spodoptera litura and S. exigua). The DNA sequences of Spodoptera frugiperda were inferred from the VCF and those of outgroup species were downloaded from the NCBI homepage. Then, multiple sequence alignment was generated using the muscle software. The neighbour joining tree was reconstructed using MEGA software with 1,000 times of bootstrapping.