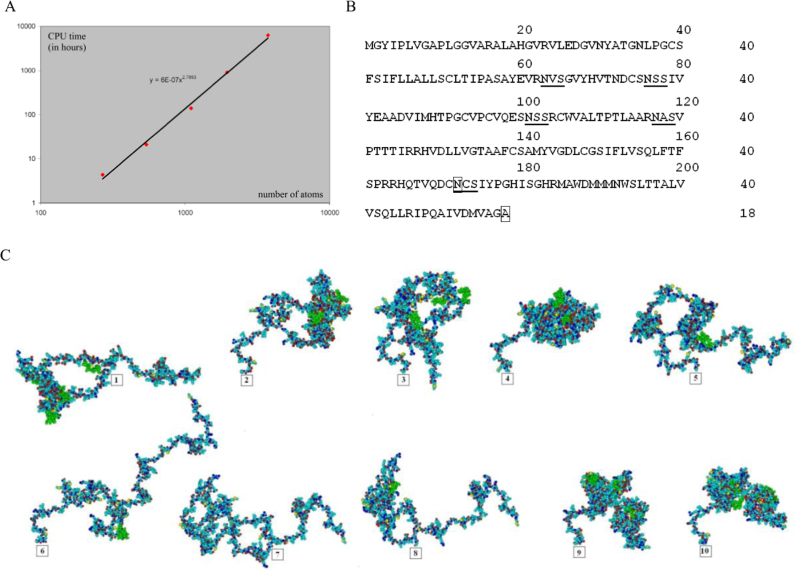
Fig. 7.
A, Dependency between the computer processing time and the number of atoms in the E1 molecule required for the full calculation of the optimum geometry of the molecule. On abscissa, the number of atoms; on ordinate, CPU time in hours. Three-dimensional structure of mutant HCV E1 protein molecules: B, Amino acid sequence of wildtype HCV E1 protein with 218 aa. The glycan attachment sites are underlined.  , the first amino-acid residue protruding from the membrane.
, the first amino-acid residue protruding from the membrane.  , the closest to the membrane asparagine to which glycan is attached; C, Computer models of mutant E1 glycoprotein variants with the disrupted glycosylation sites (N1—N6): 1, N1; 2, N5; 3, N1, N5; 4, N2, N3, and N4; 5, N2, N3, N4, and N5; 6, N1, N2, N3, and N5; 7, N1—N5; 8, N1, N2, N3, and N4; 9, with the additional glycosylation site N6; 10, initial variant of wildtype E1;
, the closest to the membrane asparagine to which glycan is attached; C, Computer models of mutant E1 glycoprotein variants with the disrupted glycosylation sites (N1—N6): 1, N1; 2, N5; 3, N1, N5; 4, N2, N3, and N4; 5, N2, N3, N4, and N5; 6, N1, N2, N3, and N5; 7, N1—N5; 8, N1, N2, N3, and N4; 9, with the additional glycosylation site N6; 10, initial variant of wildtype E1;  –
–  , the E1 membrane-anchoring site. The E1 protein is oriented so that the
, the E1 membrane-anchoring site. The E1 protein is oriented so that the  locates at the origin of the axis Oy, and
locates at the origin of the axis Oy, and  lies in the picture plane or in the plane Oxy. The scale of each protein variant was adjusted so that the sizes of all carbon atoms (light blue), or nitrogen (blue), or oxygen (red) or sulfur (yellow) were the same for all ten variants. Hydrogen is not shown. Glycans are in green color.
lies in the picture plane or in the plane Oxy. The scale of each protein variant was adjusted so that the sizes of all carbon atoms (light blue), or nitrogen (blue), or oxygen (red) or sulfur (yellow) were the same for all ten variants. Hydrogen is not shown. Glycans are in green color.