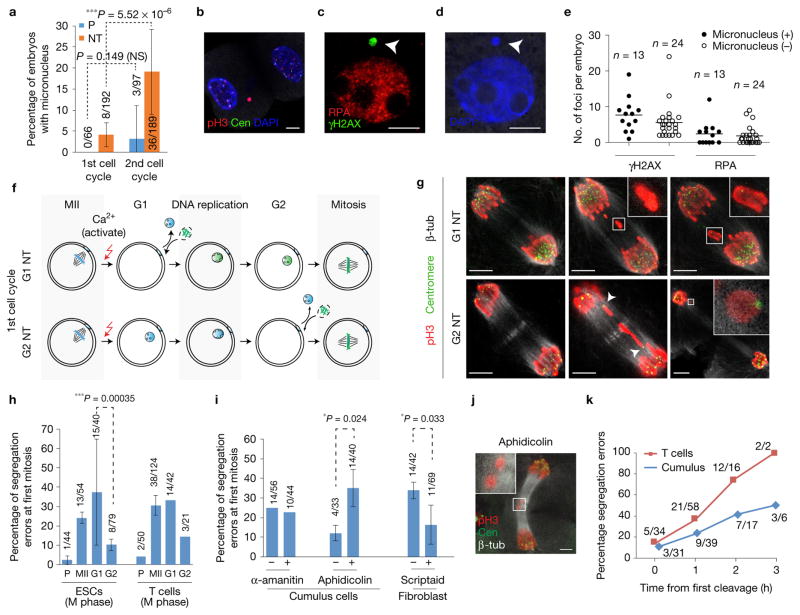
Figure 3.
Mitotic segregation defects due to DNA damage arising from progression through DNA replication. (a) Percentage of embryos with micronuclei at the first and second cell cycle. n = 66 (parthenotes (P), 1st cell cycle), 192 (NT, 1st cell cycle), 97 (parthenotes, 2nd cell cycle), 189 (NT, 2nd cell cycle) embryos. The bars represent the mean ± s.d., data are pooled from 3 independent experiments. ***P < 0.005; NS, not significant (χ2 test). (b–d) Micronucleus (arrowheads) with DNA damage in blastomeres at the interphase of the second cell cycle, 28 h post oocyte activation. (e) Quantification of DNA damage in embryos with and without micronuclei at 28 h postactivation. Lines represent the median number of foci per embryo. n=13 (micronucleus (+)) and n=24 (micronucleus (−)) embryos. (f) Schematic of NT in G1 (1 h postactivation) or G2 (13 h postactivation) using mitotic donor cells. G2 NT embryos bypass S phase, undergoing mitosis without DNA replication in the oocyte, in contrast to G1 NT embryos. Oocytes were activated with ionomycin, puromycin, cytochalasin B and 6-DMAP. (g) Representative mitotic segregation defects in G1 and G2 NT embryos. Presence of centromeres in lagging chromosomes in G2 NT embryos (arrowheads and inset), in contrast to centromere-negative fragments in G1 NT embryos (insets). (h) Frequency of mitotic segregation errors in parthenotes (P) and NT embryos where mitotic donor nuclei from ESCs or T cells are transferred into MII, G1 or G2 oocytes. Activation was performed with DMAP. The bars represent the mean ± s.d. from n = 54 (MII oocytes, ESCs), 40 (G1 oocytes, ESCs), 79 (G2 oocytes, ESCs), 124 (MII oocytes, T cells), 42 (G1 oocytes, T cells) and 21 (G2 oocytes, T cells) embryos. ***P < 0.005 (χ2 test). (i) Frequency of segregation errors at the first mitosis in NT embryos after treatment with α-amanitin (RNA polymerase II and III inhibitor), aphidicolin (DNA polymerase inhibitor) and scriptaid (HDAC inhibitor). The bars represent the mean ± s.d. from n=56(−α-amanitin), 44 (+α-amanitin), 33 (−aphidicolin), 40 (+aphidicolin), 42 (−scriptaid) and 69 (+scriptaid) embryos. *P < 0.05; NS, not significant (χ2 test). (j) Representative image of chromosome segregation defects (acentric fragments, inset) at the first mitosis in NT embryos treated with 0.025 μM aphidicolin. (k) Frequency of mitotic segregation defects as a function of timing of the first cleavage (the first cleavage within the cohort of NT embryos is set as time = 0 h). T-cell NT: n = 34 (0 h), 58 (1 h), 16 (2 h) and 2 (3 h); cumulus cell NT: n=31 (0 h), 39 (1 h), 17 (2 h) and 6 (3 h) embryos. Scale bars, 5 μm.