Abstract
Tooth development requires proliferation, differentiation, and specific migration of dental epithelial cells, through well-organized signaling interactions with mesenchymal cells. Recently, it has been reported that leucine-rich repeat-containing G protein coupled receptor 4 (LGR4), the receptor of R-spondins, is expressed in many epithelial cells in various organs and tissues and is essential for organ development and stem cell maintenance. Here, we report that LGR4 contributes to the sequential development of molars in mice. LGR4 expression in dental epithelium was detected in SOX2+ cells in the posterior end of the second molar (M2) and the early tooth germ of the third molar (M3). In keratinocyte-specific Lgr4-deficient mice (Lgr4K5 KO), the developmental defect became obvious by postnatal day 14 (P14) in M3. Lgr4K5 KO adult mice showed complete absence or the dwarfed form of M3. In M3 development in Lgr4K5 KO mice, at Wnt/β-catenin signal activity was down-regulated in the dental epithelium at P3, as indicated by lymphoid enhancer-binding factor-1 (LEF1) expression. We also confirmed the decrease, in dental epithelium of Lgr4K5 KO mice, of the number of SOX2+ cells and the arrest of cell proliferation at P7, and observed abnormal differentiation at P14. Our data demonstrated that LGR4 controls the sequential development of molars by maintaining SOX2+ cells in the dental epithelium, which have the ability to form normal molars.
Keywords: LGR4, Molar development, SOX2, Dental epithelial stem cell
Highlights
-
•
LGR4 expression was observed in the dental epithelium after birth and moved posteriorly during molar development.
-
•
Keratin5-Cre Tg specific deletion of Lgr4 impaired the development of the third molar.
-
•
LGR4 maintained SOX2 positive and proliferative cells in the dental epithelium of molars.
1. Introduction
Teeth are essential organs required to masticate food for efficient digestion. Mammals, including humans and mice, have three molars at the back of the jaws. These three molars are known to be formed by sequential developmental process. Tooth germs of the second (M2) and third molars (M3) are formed by invagination of the tooth epithelia of the first molar (M1) [1], [2]. Recently, it has been reported that dental epithelial stem cells (DESCs) transiently reside in the cervical loop (CL) of M1 [3]. Additionally, SOX2 expression, which is a marker of DESCs in M1 [3], is associated with sequential development directed to the posterior position [1]. Marcia Gaete et al. reported that epithelium of the anterior-molar tail could grow by the posterior movement of epithelial cells, followed by infolding and stratification involving a population of SOX2+/SOX9+ cells [4].
DESCs in incisors are regulated by various networks including bone morphogenetic protein (BMP), fibroblast growth factor (FGF), transforming growth factor-β (TGF-β), Notch, and Wnt [5], [6]. Wnt/β-catenin signaling is activated in dental mesenchyme, but not in the epithelium of incisor at developmental stage, and indirectly regulates Lgr5+ DESC in dental epithelium [7], [8]. Excess activation of Wnt/β-catenin signaling in the dental epithelium of incisor caused increased differentiation of DESCs [6]. In incisor, the activation of Wnt/β-catenin signaling in the dental mesenchyme and suppression in the dental epithelium is important for maintenance of DESC. However, the mechanism by which DESCs are regulated during the sequential development of molars and their effect on the development of posterior molars are unclear.
In mice, tooth development begins at around embryonic day 11.5 (E11.5) with thickening of the oral epithelium. The epithelium invaginates into the mesenchyme to form a bud by E13.5, and then forms a cap. By E16.5, the tooth germ forms a bell. During the bell stage, the epithelium adjacent to the mesenchyme differentiates into ameloblasts to produce enamel, and the mesenchyme differentiates into odontoblasts to produce dentin. This is the mechanism by which tooth germs develop through epithelial-mesenchymal interactions [9]. Similar interactions between the epithelium and mesenchyme also regulate development of the lungs, kidneys, hair follicles, and mammary glands [10], [11], [12], [13]. In addition, it is important to maintain the undifferentiated state of the epithelial cells for the development of the kidneys and mammary glands [14], [15].
In these organs, leucine-rich repeat-containing G protein coupled receptor 4 (LGR4) regulates the undifferentiated state of the epithelial cells [16], [17], and contributes to organ development. LGR4 interacts with the cell-surface transmembrane E3 ubiquitin ligase, zinc and ring finger 3 (ZNRF3), through binding to R-spondins, which are the ligands of LGR4, and clears ZNRF3 from the cell surface. This results in the inhibition of the degradation of Frizzled, a Wnt receptor, and subsequently, Wnt/β-catenin signaling is enhanced [18], [19], [20]. LGR4 is widely expressed in stem cells and proliferative components of many epithelial tissues: hair follicle, kidney, and intestine [21], [22]. When cultured ex vivo, LGR4-deficient crypts or progenitors, show marked down-regulation of stem-cell markers and Wnt target genes and die rapidly [23]. In the kidney development, LGR4 functions in maintaining the ureteric bud in an undifferentiated state [16]. Thus, LGR4 plays an important role in maintaining stem cells or undifferentiated cells in epithelial tissues. In this study, using Lgr4K5 KO mice, we demonstrated that LGR4 contributes to sequential molar development.
2. Material and methods
2.1. Animals
Lgr4EGFP-IRES-CreERT2/+ mice were kindly provided by H.C. [24]. The generation of Lgr4K5 Ctrl and Lgr4K5 KO was described previously [25]. We used mice for the analysis at the neonatal stage without distinction of sex. The care and use of mice in this study was approved by the Institutional Animal Care and Use Committee of Tohoku University.
2.2. Histological and immunohistochemical analyses
For molar histology, mice heads were fixed in 4% paraformaldehyde (PFA), and decalcified in 10% ethylenediaminetetraacetic acid (EDTA) for 1–2 weeks. After dehydration, the samples were embedded in paraffin, and the paraffin blocks were sectioned at 5 µm thickness and stained with hematoxylin-eosin (H&E). For the frozen sections, the samples were embedded in Optimal Cutting Temperature compound, and the frozen blocks were sectioned at 5 µm thickness. The immunological staining protocol used the Alexa 488-labeled goat anti-rabbit IgG antibody (Invitrogen, A-11034, 1:200), the Alexa 488-labeled goat anti-mouse IgG antibody (Invitrogen, A-11001, 1:200), the Alexa 594-labeled goat anti-mouse IgG antibody (Invitrogen, A-11005, 1:200), the peroxidase-labeled goat anti-rabbit IgG antibody (Vector Laboratories, PI-1000, 1:200), and the avidin-biotinylated enzyme complex. The paraffin-embedded sections and the frozen sections of the heads were analyzed with the following antibodies: rabbit polyclonal antibodies against green fluorescent protein (GFP; MBL, 598, 1:200), mouse monoclonal antibodies against E-cadherin (BD Biosciences, 610182, 1:500), mouse monoclonal antibodies against SOX2 (Abcam, ab79351, 1:200), rabbit polyclonal antibodies against LEF1 (Cell Signaling Technologies, #2230S, 1:100), rabbit monoclonal antibodies against SOX2 (Abcam, ab92494, 1:200), rabbit polyclonal antibodies against Ki67 (Novus Biologicals, NB110-89717, 1:200), mouse monoclonal antibodies against bromodeoxyuridine (BrdU; Roche, 11,170,376,001, 1:200), and rabbit polyclonal antibodies against Amelogenin (Santa Cruz, sc-32892, 1:100). These antibodies were diluted with 5% normal goat serum or the Mouse on Mouse immunodetection kit (Vector Laboratories) in Tris-buffered saline (TBS) or phosphate buffer (PB) and applied to the sections, which were then incubated overnight at 4 °C.
2.3. 5-Bromo-2′-deoxyuridine (BrdU) administration
BrdU (Roche) was dissolved in phosphate buffered saline (PBS) at 5 mg/ml and injected intraperitoneally (50 µg/g body weight).
2.4. Statistics
The results of the experiments are expressed as the means±SEM. ANOVA and Student's t-test were used for statistical analysis of the results, and a value of P<0.05 was considered statistically significant.
3. Results
3.1. Lgr4 is expressed in the bud epithelium of M1 at the embryonic stage, and its expression shifts to the posterior molar at the postnatal stage
During M1 development, Lgr4 expression in the dental epithelium from E10.5 to E13.5, and in the collar of the tooth and the outer enamel epithelium at E14.5 has been reported [26]. However, the Lgr4 expression pattern during M2 or M3 development is not known.
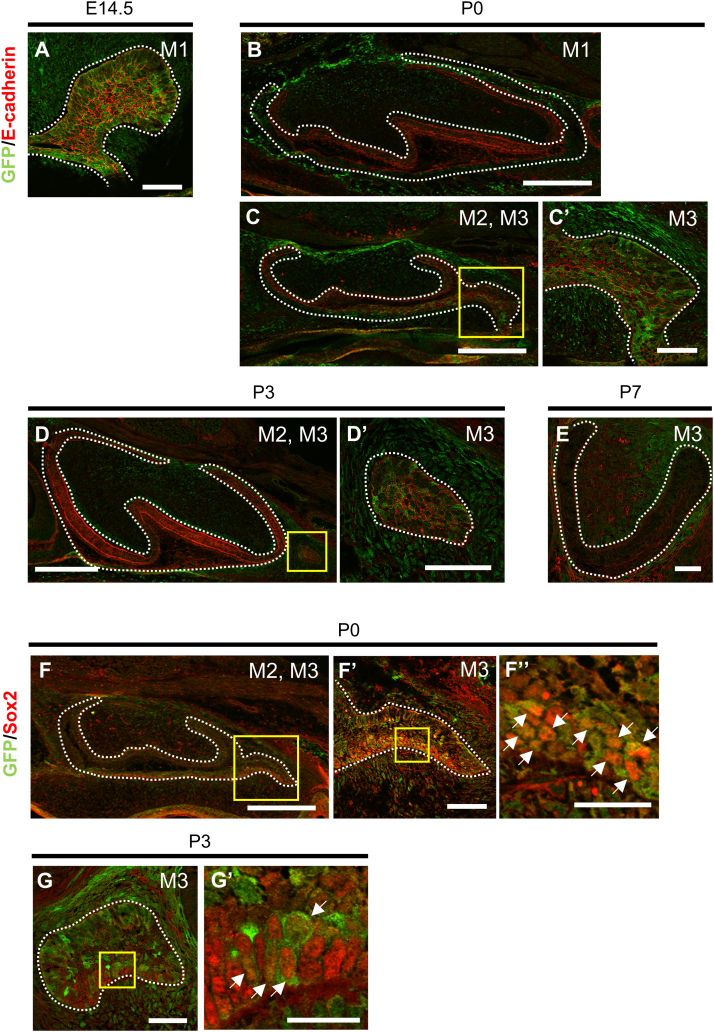
In this study, we analyzed the expression pattern of LGR4 during molar development using Lgr4EGFP-IRES-CreERT2/+ mice, which express EGFP (and Cre ERT2) inserted in the Lgr4 locus. Using immunostaining, we detected EGFP colocalized with E-cadherin, an epithelial marker. Strong expression of EGFP was detected in the dental epithelium of M1 at E14.5, indicating that LGR4 was expressed there (Fig. 1A). This result is consistent with those of previous reports [26]. At postnatal day 0 (P0), the EGFP signal was not detectable in the M1 tooth epithelium region (Fig. 1B), but was observed in the collar of M2, and in the bud of M3, which formed from the posterior part of M2 (Fig. 1C, C′). At P3, EGFP was only expressed in the epithelium of M3 (Fig. 1D, D′). No expression was observed in the dental epithelium at P7, by which time M3 develops to the bell stage (Fig. 1E). We found that LGR4 expression shifted to the posterior epithelium, accompanied by the sequential formation of molars from M1 to M3. Next, we performed double immunostaining for EGFP and SOX2, a DESC marker in incisors [27]. The nuclear signal of SOX2 is only detected in the posterior end and in the outer enamel epithelium of M2 and in M3 bud. The expression of EGFP was closely associated with SOX2 expression in the dental epithelium of M2 and M3 at P0 (Fig. 1F-F′′). At P3, EGFP was expressed in a part of SOX2+ epithelial cells (Fig. 1G, G′). Taken together, these results suggest that LGR4 is expressed in the SOX2+ epithelial cells during sequential molar development.
Fig. 1.
Lgr4 is expressed in the bud epithelium of M1 at the embryonic stage, and its expression shifts to the posterior molar at the postnatal stage. (A-E) Immunofluorescence analysis for GFP (green) and E-cadherin (red) of tooth germ in Lgr4EGFP-IRES-CreERT2/+ mice in coronal (A) and sagittal sections (B-E). (F and G) Immunofluorescence analysis for GFP (green) and SOX2 (red) in Lgr4EGFP-IRES-CreERT2/+mice in sagittal sections. Arrows indicate GFP+/SOX2+ cells. Boxed areas in (C), (D), (F), (F′), and (G) are shown magnified in (C′), (D′), (F’), (F′′), and (G′) respectively. Dot-lines indicate dental epithelium. The sample size (n) for each experimental group/condition was 3. Three different individuals were used in the experiment. All experiments were performed in duplicate to confirm the repeatability. Scale bars represent 50 µm (A, C′, D′, E, F′, and G), 200 µm (B-D, and F), and 20 µm (F′′ and G′).
3.2. Keratin5-Cre Tg specific deletion of Lgr4 affects the development of M3
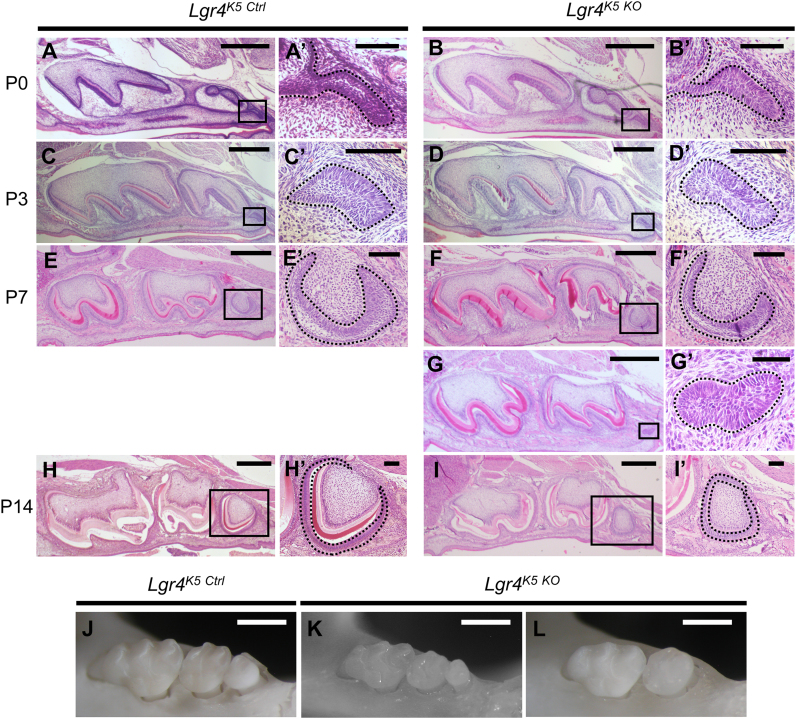
To study the role of LGR4 in molar development, we first analyzed the mouse model with targeted deletion of Lgr4 gene. We histologically analyzed Lgr4 null mice (Lgr4-/-). These mice exhibited neonatal lethality due to renal hypoplasia and impaired erythropoiesis [28], [29]. However, they did not show any aberration in the tooth germ of M1 or M2 at P0 (Fig. S1A). To analyze the role of LGR4 in molar development, we generated conditional knock-out mice crossed with Keratin5-Cre (K5-Cre) transgenic (Tg) mice. One of the proteins of the cytokeratin family, Keratin5, is expressed in various epithelial cells including the oral epithelium [30]. Keratin5 promoter has been used in some mouse models for over expression or gene deletion in tooth germ epithelial cells [31]. Lgr4 conditional knock-out mice with K5-Cre Tg mice (Lgr4K5 KO) circumvented neonatal lethality and kidney abnormalities [25]. The molars in Lgr4K5 KO mice morphologically developed to their normal structure, similar to in Lgr4K5 Ctrl mice, until P3 when M3 developed to the bud stage (Fig. 2A-D, A′-D′). At P7, all Lgr4K5 Ctrl and most Lgr4K5 KO mice showed development to the cap stage in M3 (Fig. 2E, F, E′, F′), but M3 in only one of the Lgr4K5 KO mice formed a bud (Fig. 2G, G′). In addition, M3 in Lgr4K5 Ctrl formed “the late bell” structure and developed the layer of enamel or dentin at P14 (Fig. 2H, H′); however, M3 in Lgr4K5 KO formed “the early bell” structure and no layer of hard tissue was observed at P14 (Fig. 2I, I′). Additionally, at P14, there was individual with no M3 germ although we observed sequential sections (data not shown). Thus, we confirmed the abnormal development of M3 in Lgr4K5 KO mice during the period from P7 to P14.
Fig. 2.
Keratin5-Cre Tg specific deletion of Lgr4 affects the development of M3. (A-I) H&E staining of Lgr4K5 Ctrl and Lgr4K5 KO mice in sagittal sections. Boxed areas in (A)-(I) are shown magnified in (A′)-(I′), respectively. Dot-lines indicate dental epithelium of M3. (J-L) Macroscopic views of maxillary molars in male Lgr4K5 Ctrl and Lgr4K5 KO mice at 10 weeks age. The sample size (n) for the experimental groups/conditions is 3 (A-I), and 15 (J-L). Different individuals were used in the experiment. All experiments were performed in duplicate to confirm the repeatability. Scale bars represent 500 µm (A-I), 50 µm (A′-I′), and 1 mm (J-L).
Next, we analyzed molars in male Lgr4K5 Ctrl or Lgr4K5 KO mice at postnatal 10 weeks. Some of the Lgr4K5 KO mice showed the dwarfed form of upper M3 (9/15; 60%, Fig. 2K) and the others showed its complete absence (5/15; 33%, Fig. 2L). Only in one out of the 15 Lgr4K5 KO mice analyzed, M3 developed to its normal structure (1/15, 7%) similar to that of Lgr4K5 Ctrl (Fig. 2J). This abnormality was also observed in the lower M3 (Fig. S1B). The size of M1 and M2 in Lgr4K5 KO mice was slightly, but significantly, decreased compared to that of normal M1 and M2. However, the severest abnormality was observed in M3 (Fig. S1C-F). We found that in Lgr4K5 KO mice, the development of M3 was abnormal at the early stage, and it was completely absent or dwarfed at adulthood.
3.3. Lgr4 deletion disrupts the differentiation of dental epithelium into ameloblasts
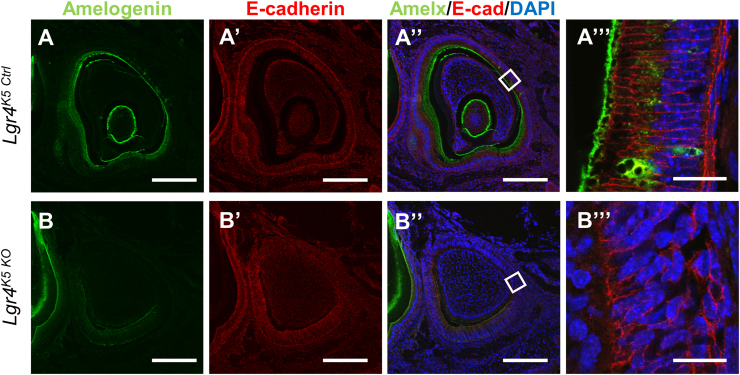
In the tooth development, dental epithelium differentiates into ameloblasts to secrete enamel after the bell stage. Amelogenesis is the process of differentiation of dental epithelial cells into ameloblasts, which secrete enamel. To secrete enamel, mature ameloblast forms columnar structure with the localization of nuclei near the stellate reticulum side [32]. Fig. 2 showed that Lgr4 deletion caused enamel defect in M3 at P14. Therefore, we examined the differentiation of M3 epithelium in Lgr4K5 KO mice. These mice showed abnormal cell polarity and decreased expression of Amelogenin, the enamel matrix protein, in M3 ameloblasts at P14 (Fig. 3A-A′′′, B-B′′′). This result indicates that LGR4 was involved in the differentiation of dental epithelial cells into ameloblasts in M3.
Fig. 3.
Lgr4 deletion disrupts the differentiation of dental epithelium into ameloblasts (A and B) Immunofluorescence analysis for Amelogenin (green) and E-cadherin (red) of M3 tooth germ in Lgr4K5 Ctrl and Lgr4K5 KO mice in sagittal sections. Counterstain is DAPI. Boxed areas in (A′′) and (B′′) are shown magnified in (A′′′) and (B′′′), respectively. Amelx - Amelogenin; E-cad - E-cadherin The sample size (n) for each experimental group/condition is 3. Three different individuals were used in the experiment. All experiments were performed in duplicate to confirm the repeatability. Scale bars represent 200 µm (A-A′′ and B-B′′) and 20 µm (A′′′ and B′′′).
3.4. LGR4 promotes Wnt/β-catenin signaling in M3 development
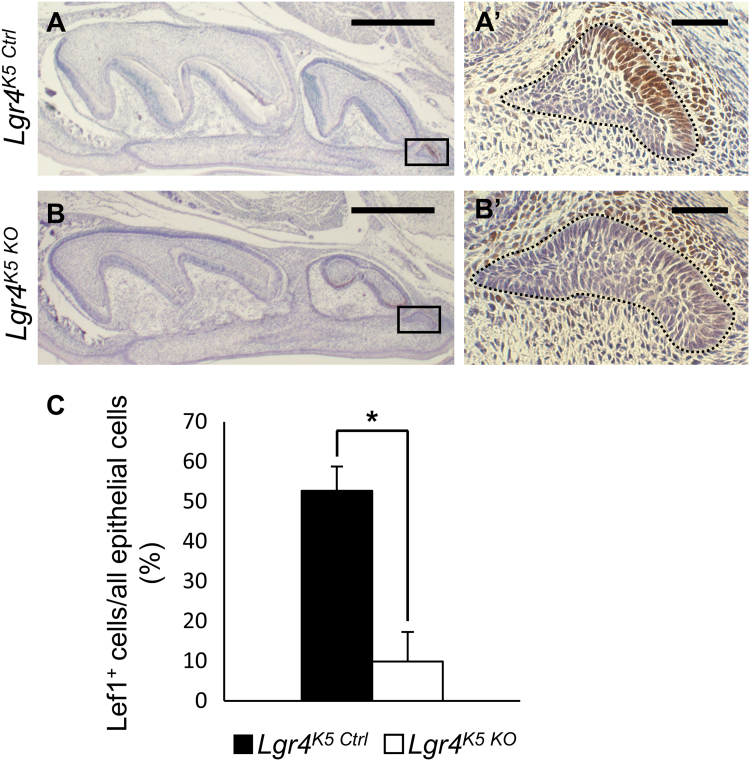
LGR4 enhances Wnt/β-catenin signaling by binding its ligand, R-spondin [19], [20]. Wnt/β-catenin signaling plays an essential role in the early stage of molar development. It was reported that β-catenin or LEF1 knock-out mice showed developmental arrest of M1 at bud stage [33], [34], [35]. Therefore, we evaluated Wnt/β-catenin signaling activity in M3 development by immunostaining for LEF1, a downstream target of Wnt signaling. Lgr4K5 Ctrl mice showed strong expression of LEF1 in M3 dental epithelium and mesenchyme at P3 (Fig. 4A, A′). This expression pattern was similar to that of M1 bud stage [33]. However, LEF1 was hardly detected in M3 dental epithelium of Lgr4K5 KO (Fig. 4B, B′), and the percentage of LEF1+ epithelial cells was significantly decreased (Fig. 4C). These results indicate that LGR4 enhances Wnt/β-catenin signaling in M3 dental epithelium.
Fig. 4.
Lgr4 promotes Wnt/β-catenin signaling in M3 development (A and B) Immunohistochemistry of LEF1 in Lgr4K5 Ctrl and Lgr4K5 KO mice in sagittal sections at P3. Counterstain is hematoxylin. Dot-lines indicate dental epithelium of M3. Boxed areas in (A) and (B) are shown magnified in (A′) and (B′), respectively. Scale bars represent 500 µm (A and B) and 50 µm (A′ and B′). (C) The percentage of LEF1+ cells in M3 dental epithelium of Lgr4K5 Ctrl and Lgr4K5 KO mice. N=3. *p<0.05. Error bars, SEM. ANOVA and Student's t-test were used for the statistical analysis. The sample size (n) for each experimental group/condition is 3. Three different individuals were used in the experiment. All experiments were performed in duplicate to confirm repeatability.
3.5. Lgr4 deletion decreases the SOX2 positive and proliferative cells in the dental epithelium during molar development
LGR4 activates SOX2 expression and regulates the development and stem cell functions in mammary gland via Wnt/β-catenin/LEF1 signaling [17]. Wnt signal regulates proliferation and differentiation of SOX2+ cells during cochlea development [36]. In the molar development, SOX2+ cells contribute to sequential molar development [1], [4]. In the present study, we examined the expression of SOX2, using Lgr4K5 KO mice.
We confirmed the expression of SOX2 in the dental epithelium of M3, dental cord, and dental lamina connecting M1 to M2, in both Lgr4K5 Ctrl and Lgr4K5 KO mice until P3 (Fig. 5A-D, A′-D′). At P7, SOX2 was localized in the CL of M3 in Lgr4K5 Ctrl (Fig. 5E-E′′). In contrast, no expression of SOX2 was observed in the M3 dental epithelium of Lgr4K5 KO (Fig. 5F-F′′). At P14, SOX2 was expressed in the M3 dental cord of Lgr4K5 Ctrl, but not in the CL (Fig. 5G, G′). No expression of SOX2 was observed in M3 dental epithelium of Lgr4K5 KO (Fig. 5H, H′). In addition, the percentage of SOX2+ cells in M3 epithelium of Lgr4K5 KO was comparable with that of Lgr4K5 Ctrl at P3, but significantly decreased at P7 (Fig. 5I, J). Our results strongly demonstrate that the loss of Lgr4 causes a decrease in the number of SOX2+ cells in the M3 dental epithelium during early developmental stage.
Fig. 5.
Lgr4 deletion decreased the SOX2+ cells in the dental epithelium during molar development. (A-H) Immunohistochemistry of SOX2 in Lgr4K5 Ctrl and Lgr4K5 KO mice in sagittal sections at P0, P3, P7, and P14. Counterstain is hematoxylin. Dot-lines indicate dental epithelium of M3. Boxed areas in (A)-(H), (E′), and (F′) are shown magnified in (A′)-(H′), (E′′), and (F′′), respectively. Scale bars represent 500 µm (A-H) and 50 µm (A′-H′, E′′, and F′′). (I and J) The percentage of SOX2+ cells in M3 dental epithelium of Lgr4K5 Ctrl and Lgr4K5 KO mice at P3 and P7. N=3. *p<0.05. Error bars, SEM. ANOVA and Student's t-test were used for the statistical analysis. The sample size (n) for each experimental group/condition is 3. All experiments were performed in duplicate to confirm the repeatability.
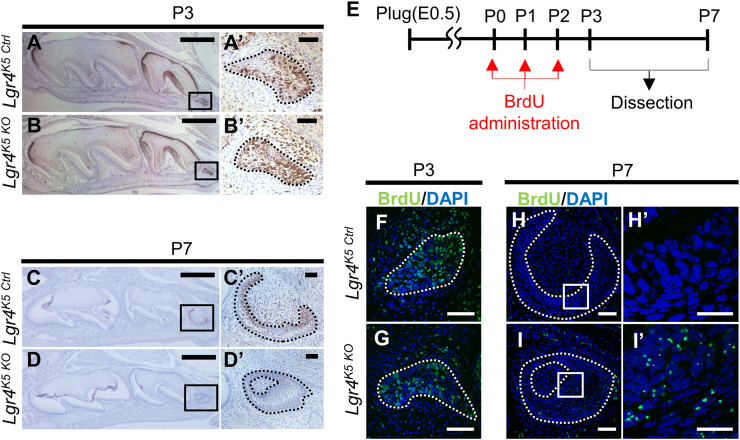
SOX2+ cells in the dental epithelium have high proliferative activity in the M1 development [3]. Therefore, we expected that abnormal cell proliferation occurred in Lgr4K5 KO mice at P7. We performed immunostaining for antigen Ki67, a typical cell proliferation marker. At P3, there was no difference in M3 between Lgr4K5 Ctrl (Fig. 6A, A′) and Lgr4K5 KO mice (Fig. 6B, B′) in the expression of Ki67. However, the expression of Ki67 in the dental epithelium of M3 of Lgr4K5 KO was significantly decreased at P7 (Fig. 6D, D′) compared to that in Lgr4K5 Ctrl (Fig. 6C, C′). Therefore, we hypothesized that epithelial cell proliferation was inhibited in M3 of Lgr4K5 KO from P3 to P7. To determine the proliferative rate of these cells, we conducted pulse chase analysis with BrdU. BrdU, an analog of thymidine, is incorporated into the nuclei of cells during DNA synthesis, and the cells labeled with BrdU can be detected by the anti-BrdU antibody. BrdU density in DNA finally decreases to below the detectable level following repeated cell division. However, the cells with slow cell cycle can be detected even after the lapse of a long duration after BrdU administration [37], [38], [39], [40].
Fig. 6.
LGR4 regulated the cell proliferation of the dental epithelium in M3. (A-D) Immunohistochemistry of Ki67 in Lgr4K5 Ctrl and Lgr4K5 KO mice in sagittal sections at P3 and P7. Counterstain is hematoxylin. (E) Timing of BrdU administration and sample harvest in Lgr4K5 Ctrl and Lgr4K5 KO mice. (F-I) Immunofluorescence analysis for BrdU in Lgr4K5 Ctrl and Lgr4K5 KO mice in sagittal sections at P3 and P7. Counterstain is DAPI. Dot-lines indicate dental epithelium of M3. Boxed areas in (A)-(D), (H), and (I) are shown magnified in (A′)-(D′), (H′), and (I′), respectively. The sample size (n) for each experimental group/condition is 3. Three different individuals were used in the experiment. All experiments were performed in duplicate to confirm the repeatability. Scale bars represent 500 µm (A-D), 50 µm (A′-D′, F-I), and 20 µm (H′ and I′).
We administrated BrdU intraperitoneally to Lgr4K5 KO mice from P0 to P2, dissected these mice at P3 or P7, and detected cells with BrdU by immunostaining (Fig. 6E). At P3, BrdU was detected in the dental epithelium of M3 of Lgr4K5 Ctrl and Lgr4K5 KO (Fig. 6F, G). At P7, BrdU+ cells were detected in M3 of Lgr4K5 KO (Fig. 6I, I′), but no signal was detected in Lgr4K5 Ctrl (Fig. 6H, H′). In Lgr4K5 Ctrl mice, BrdU density in DNA may have been below the detectable level because the epithelial cells proliferated actively. In contrast, BrdU was detected at P7 in Lgr4K5 KO mice suggesting that cell proliferation is inhibited in the dental epithelium of M3 in Lgr4K5 KO in the period from P3 to P7. Our data demonstrates that Lgr4 deletion diminishes the cells that actively proliferate in the dental epithelium.
4. Discussion
Three molars develop sequentially from the first tooth germ through invagination of the dental epithelium to form the posterior tooth germ of the next molar. In this report, we clarified the role of LGR4 in sequential molar development using Lgr4K5 KO mice, which showed an abnormal morphology of the tooth germ and altered gene expression in M3 during the neonatal stage and loss of M3 loss or dwarfed M3 in adults (Table 1).
Table 1.
The summary of time points and differences between Lgr4K5 Ctrl and Lgr4K5 KO.
| Lgr4K5 Ctrl | Lgr4K5 KO | |
|---|---|---|
| P0 | M3 is formed by invagination of M2 epithelium. | |
| P3 |
|
|
| P7 |
|
|
| P14 |
|
|
| Adult | Molar development is completed. | A dwarfed form or the complete absence of M3 |
In the early fetal stage, SOX2+ cells exist in the dental epithelium of M1 [1]. SOX2 expression is translocated to the posterior epithelium of M1 concomitantly with the development and contributes to epithelial cell lineage in M2 or M3 [1]. Using Lgr4EGFP-IRES-CreERT2/+ mice, we found that the expression of LGR4 was observed in some of the SOX2+ cells and translocated to the posterior epithelium as molars sequentially developed from M1 to M3. This expression pattern and its transition appear to be similar to that observed in SOX2+ cells. These results suggest that SOX2+ cells as well as other undifferentiated epithelial cells express LGR4. LGR4 maintains undifferentiated cells in the kidneys, mammary glands, and intestines and contributes to normal development and homeostasis [16], [17].
To study the role of LGR4 in molar development, we used the loss of function approach with gene-deleted mouse models. Interestingly, Ki67+ proliferative cells were not decreased at P3, but clearly decreased at P7 in Lgr4K5 KO mice. In addition, we administrated BrdU to newborn pups of Lgr4K5 Ctrl during the period from P0 to P2, and BrdU was undetectable at P7; however it persisted in M3 of Lgr4K5 KO mice at P7 (Fig. 6). These data demonstrate that the epithelial cells of Lgr4K5 KO gradually lose their division potential during the M3 tooth germ development phase. Moreover, at P7, SOX2+ cells in CL were not retained in Lgr4K5 KO mice. These data suggest that LGR4 regulates the sequential development of molars by maintaining SOX2+ cells, which have a higher potential to proliferate.
The role of Wnt/β-catenin signaling in the regulation of sequential development in molars has been unclear. We showed that Lgr4 ablation caused the suppression of Wnt signal activity in M3, using the experimental model of Lgr4K5 KO mice. Our data indicate that LGR4 is involved in the activation of Wnt signaling during posterior molar development. However, Wnt signal is down-regulated during maintenance of SOX2+ DESC in incisors. The activation of Wnt signal by a drug (BIO) decreases SOX2 expression and inhibits cell proliferation in CL of incisor [41]. In addition, SOX2+ DESCs transiently express the Wnt inhibitor Sfrp5 and contribute to all epithelial cell lineages of the tooth [27]. These observations suggest that Wnt signal plays different roles in DESC regulation: the maintenance of SOX2+ cells in the molar and the promotion of differentiation in incisor. Additionally, decreased LEF1 expression in Lgr4K5 KO mice was observed at P3, whereas the number of SOX2+ cells and the proliferative ability were decreased after P7 (Table 1). This temporal difference in the timing of the alteration between SOX2 and LEF1 expression might reflects the indirect relationship between Wnt signaling and SOX2 expression. However, it still remains unclear whether Wnt signaling regulates SOX2 expression per this report. Further experiments will be required to investigate the involvement between Wnt signaling and SOX2+ epithelial cells.
In the M3 ameloblast of Lgr4K5 KO mice, the cell polarity was disrupted with abnormal localization and the enamel secretion was inhibited at P14 (Fig. 3). This result indicates that Lgr4K5 KO mice lack mature ameloblasts in the tooth germ of M3. However, Lgr4 was only expressed during the early developmental stage of M3, but not expressed after P7 at which time ameloblast differentiation and maturation occurs. Therefore, LGR4 does not directly regulate ameloblast differentiation. These data suggested that LGR4 maintains the germ cells of ameloblast in the dental epithelium during early developmental stage. The maintenance of dental epithelium during developmental phase by Lgr4 probably influences the characteristics of M3, such as structure, existence, and stability to jaws in adult mice.
There was a variation in the phenotypes of M3 within the experimental group of Lgr4K5 KO mice: small structure, dislocation from jaws, and complete loss. These variations were observed during the neonatal stage, where some Lgr4K5 KO mice showed an abnormal M3 germ that failed to form a cap at P7 (Fig. 2G). At P14, in addition to individuals with defective enamel, the M3 germ was completely lost in some Lgr4K5 KO mice (data not shown). These results suggest the possibility that the progenitor is disrupted in Lgr4K5 KO mice with severe phenotypes at the adult stage.
The full length of M1 and M2 was slightly decreased in Lgr4K5 KO mice at the adult stage (Fig. S1C-F). In contrast, the tooth germ of M1 and M2 was normally developed in Lgr4K5 KO mice or in Lgr4-/- mice at P0, and no critical differences were observed by morphological analysis (Fig. 2A, B and Fig. S1A, B). We presume that the subtle abnormalities in the dental epithelial progenitor at the early developmental stage, which we were unable to determine in this study, are caused by Lgr4 deletion, and they finally caused little abnormality in M1 and M2 at the adult stage. These subtle abnormalities, for example, the decrease of division frequency or the reduction of the period for maintaining the undifferentiated state, might affect the size of the tooth germ or the differentiation potential to ameloblasts in M1 and M2 development.
In this study, we used both male and female Lgr4K5 KO mice in the analysis at neonatal stage. At adult, male Lgr4K5 KO mice showed the dwarfed form or complete absence in M3 (Fig. 2J-L). We also performed the morphological analysis of female Lgr4K5 KO mice at adult. As a result, female showed similar phenotype as seen in male. No gender-related differences were observed in adult mice, which probably true in neonatal mice as well.
In mice, molars have no potential to grow continuously or to be replaced, and this feature is similar to that of molars in humans. Additionally, the mechanism underlying the sequential development of molars is probably similar in mice and humans. However, this mechanism has been mostly unclear. In this study, using Lgr4 deficient mice, we demonstrated for the first time that LGR4 is required for the maintenance of SOX2+ cells and their proliferation during the sequential development of molars from the anterior to posterior epithelia of the tooth germs. This study has afforded a better understanding of the maintenance mechanism of DESCs during molar development, which should accelerate the development of new approaches for studying the mechanisms underlying tooth regeneration.
Acknowledgments
We are grateful to Hans Clevers for kindly providing the Lgr4EGFP-IRES-CreERT2 mice. This study was supported by Grants-in-Aid for Scientific Research on Innovative Areas.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.bbrep.2016.08.018.
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.08.018.
Appendix A. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Appendix B. Supplementary material
Supplementary material
.
References
- 1.Juuri E., Jussila M., Seidel K., Holmes S., Wu P., Richman J., Heikinheimo K., Chuong C.M., Arnold K., Hochedlinger K., Klein O., Michon F., Thesleff I. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development. 2013;140:1424–1432. doi: 10.1242/dev.089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juuri E., Isaksson S., Jussila M., Heikinheimo K., Thesleff I. Expression of the stem cell marker, SOX2, in ameloblastoma and dental epithelium. Eur. J. Oral Sci. 2013;121:509–516. doi: 10.1111/eos.12095. [DOI] [PubMed] [Google Scholar]
- 3.Li J., Feng J., Liu Y., Ho T.V., Grimes W., Ho H.A., Park S., Wang S., Chai Y. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev. Cell. 2015;33:125–135. doi: 10.1016/j.devcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaete M., Fons J.M., Popa E.M., Chatzeli L., Tucker A.S. Epithelial topography for repetitive tooth formation. Biol. Open. 2015 doi: 10.1242/bio.013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuang-Hsien J., Mushegyan Hu, V., Klein O.D. On the cutting edge of organ renewal: Identification, regulation, and evolution of incisor stem cells. Genesis. 2014;52:79–92. doi: 10.1002/dvg.22732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang G., Zhou J., Teng Y., Xie J., Lin J., Guo X., Gao Y., He M., Yang X., Wang S. Mesenchymal TGF-beta signaling orchestrates dental epithelial stem cell homeostasis through Wnt signaling. Stem Cells. 2014;32:2939–2948. doi: 10.1002/stem.1772. [DOI] [PubMed] [Google Scholar]
- 7.Suomalainen M., Thesleff I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev. Dyn. 2010;239:364–372. doi: 10.1002/dvdy.22106. [DOI] [PubMed] [Google Scholar]
- 8.Chang J.Y., Wang C., Liu J., Huang Y., Jin C., Yang C., Hai B., Liu F., D'Souza R.N., McKeehan W.L., Wang F. Fibroblast growth factor signaling is essential for self-renewal of dental epithelial stem cells. J. Biol. Chem. 2013;288:28952–28961. doi: 10.1074/jbc.M113.506873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucker A., Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 10.Rishikaysh P., Dev K., Diaz D., Qureshi W.M., Filip S., Mokry J. Signaling involved in hair follicle morphogenesis and development. Int J. Mol. Sci. 2014;15:1647–1670. doi: 10.3390/ijms15011647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan B.L., Yingling J.M. Epithelial/mesenchymal interactions and branching morphogenesis of the lung. Curr. Opin. Genet Dev. 1998;8:481–486. doi: 10.1016/s0959-437x(98)80121-4. [DOI] [PubMed] [Google Scholar]
- 12.Kuure S., Vuolteenaho R., Vainio S. Kidney morphogenesis: cellular and molecular regulation. Mech. Dev. 2000;92:31–45. doi: 10.1016/s0925-4773(99)00323-8. [DOI] [PubMed] [Google Scholar]
- 13.Pispa J., Thesleff I. Mechanisms of ectodermal organogenesis. Dev. Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 14.Grote D., Boualia S.K., Souabni A., Merkel C., Chi X., Costantini F., Carroll T., Bouchard M. Gata3 acts downstream of beta-catenin signaling to prevent ectopic metanephric kidney induction. PLoS Genet. 2008;4:e1000316. doi: 10.1371/journal.pgen.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith G.H. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 16.Mohri Y., Oyama K., Akamatsu A., Kato S., Nishimori K. Lgr4-deficient mice showed premature differentiation of ureteric bud with reduced expression of Wnt effector Lef1 and Gata3. Dev. Dyn. 2011;240:1626–1634. doi: 10.1002/dvdy.22651. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Dong J., Li D., Lai L., Siwko S., Li Y., Liu M. Lgr4 regulates mammary gland development and stem cell activity through the pluripotency transcription factor Sox2. Stem Cells. 2013;31:1921–1931. doi: 10.1002/stem.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao H.X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., Lei H., Mickanin C., Liu D., Ruffner H., Mao X., Ma Q., Zamponi R., Bouwmeester T., Finan P.M., Kirschner M.W., Porter J.A., Serluca F.C., Cong F. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 20.Zebisch M., Jones E.Y. ZNRF3/RNF43 – a direct linkage of extracellular recognition and E3 ligase activity to modulate cell surface signalling. Prog. Biophys. Mol. Biol. 2015;118:112–118. doi: 10.1016/j.pbiomolbio.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Barker N., Tan S., Clevers H. Lgr proteins in epithelial stem cell biology. Development. 2013;140:2484–2494. doi: 10.1242/dev.083113. [DOI] [PubMed] [Google Scholar]
- 22.Van Schoore G., Mendive F., Pochet R., Vassart G. Expression pattern of the orphan receptor LGR4/GPR48 gene in the mouse. Histochem Cell Biol. 2005;124:35–50. doi: 10.1007/s00418-005-0002-3. [DOI] [PubMed] [Google Scholar]
- 23.Mustata R.C., Van Loy T., Lefort A., Libert F., Strollo S., Vassart G., Garcia M.I. Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO Rep. 2011;12:558–564. doi: 10.1038/embor.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., Stange D.E., van Es J.E., Guardavaccaro D., Schasfoort R.B., Mohri Y., Nishimori K., Mohammed S., Heck A.J., Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 25.Kato S., Mohri Y., Matsuo T., Ogawa E., Umezawa A., Okuyama R., Nishimori K. Eye-open at birth phenotype with reduced keratinocyte motility in LGR4 null mice. FEBS Lett. 2007;581:4685–4690. doi: 10.1016/j.febslet.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki M., Porntaveetus T., Kawasaki K., Oommen S., Otsuka-Tanaka Y., Hishinuma M., Nomoto T., Maeda T., Takubo K., Suda T., Sharpe P.T., Ohazama A. R-spondins/Lgrs expression in tooth development. Dev. Dyn. 2014;243:844–851. doi: 10.1002/dvdy.24124. [DOI] [PubMed] [Google Scholar]
- 27.Juuri E., Saito K., Ahtiainen L., Seidel K., Tummers M., Hochedlinger K., Klein O.D., Thesleff I., Michon F. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev. Cell. 2012;23:317–328. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato S., Matsubara M., Matsuo T., Mohri Y., Kazama I., Hatano R., Umezawa A., Nishimori K. Leucine-rich repeat-containing G protein-coupled receptor-4 (LGR4, Gpr48) is essential for renal development in mice. Nephron Exp. Nephrol. 2006;104:e63–e75. doi: 10.1159/000093999. [DOI] [PubMed] [Google Scholar]
- 29.Song H., Luo J., Luo W., Weng J., Wang Z., Li B., Li D., Liu M. Inactivation of G-protein-coupled receptor 48 (Gpr48/Lgr4) impairs definitive erythropoiesis at midgestation through down-regulation of the ATF4 signaling pathway. J. Biol. Chem. 2008;283:36687–36697. doi: 10.1074/jbc.M800721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klopcic B., Maass T., Meyer E., Lehr H.A., Metzger D., Chambon P., Mann A., Blessing M. TGF-beta superfamily signaling is essential for tooth and hair morphogenesis and differentiation. Eur. J. Cell Biol. 2007;86:781–799. doi: 10.1016/j.ejcb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Ida-Yonemochi H., Satokata I., Ohshima H., Sato T., Yokoyama M., Yamada Y., Saku T. Morphogenetic roles of perlecan in the tooth enamel organ: an analysis of overexpression using transgenic mice. Matrix Biol. 2011;30:379–388. doi: 10.1016/j.matbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Liu C., Niu Y., Zhou X., Xu X., Yang Y., Zhang Y., Zheng L. Cell cycle control, DNA damage repair, and apoptosis-related pathways control pre-ameloblasts differentiation during tooth development. BMC Genom. 2015;16:592. doi: 10.1186/s12864-015-1783-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F., Chu E.Y., Watt B., Zhang Y., Gallant N.M., Andl T., Yang S.H., Lu M.M., Piccolo S., Schmidt-Ullrich R., Taketo M.M., Morrisey E.E., Atit R., Dlugosz A.A., Millar S.E. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Lan Y., Baek J.A., Gao Y., Jiang R. Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev. Biol. 2009;334:174–185. doi: 10.1016/j.ydbio.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kratochwil K., Galceran J., Tontsch S., Roth W., Grosschedl R. FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(-/-) mice. Genes Dev. 2002;16:3173–3185. doi: 10.1101/gad.1035602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munnamalai V., Fekete D.M. Wnt signaling during cochlear development. Semin. Cell Dev. Biol. 2013;24:480–489. doi: 10.1016/j.semcdb.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson C.B., Momma S., Clarke D.L., Risling M., Lendahl U., Frisen J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- 38.Oliver J.A., Maarouf O., Cheema F.H., Martens T.P., Al-Awqati Q. The renal papilla is a niche for adult kidney stem cells. J. Clin. Invest. 2004;114:795–804. doi: 10.1172/JCI20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams D.C., Oxburgh L. The long-term label retaining population of the renal papilla arises through divergent regional growth of the kidney. Am. J. Physiol. Ren. Physiol. 2009;297:F809–F815. doi: 10.1152/ajprenal.90650.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dey-Guha I., Mukhopadhyay M., Phillips M., Westphal H. Role of ldb1 in adult intestinal homeostasis. Int J. Biol. Sci. 2009;5:686–694. doi: 10.7150/ijbs.5.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z., Balic A., Michon F., Juuri E., Thesleff I. Mesenchymal Wnt/beta-catenin signaling controls epithelial stem cell homeostasis in teeth by inhibiting the antiapoptotic effect of Fgf10. Stem Cells. 2015;33:1670–1681. doi: 10.1002/stem.1972. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material