Figure 5. Binding of the full range of CAP256 lineage Abs to CRF250 SOSIP.664 shows a gradation of sensitivity to α2,6 sialidase treatment dependent upon key HCDR2 residues.
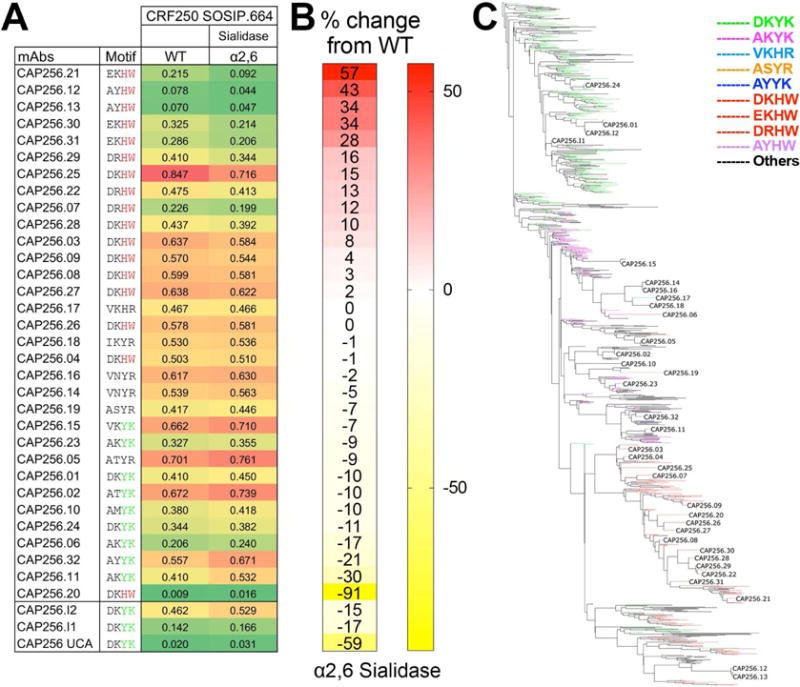
A. Octet binding of CAP256-derived mAbs to PGT145 Ab-affinity purified CRF250 SOSIP.664 trimer (WT) and α2,6 desialylated version. The CAP256 mAbs were captured on an anti-human IgG-Fc sensor and then the CRF250 trimer forms (100nM) flowed over. The binding response (nanometers (nm)) to WT and the α2,6 desialylated trimers is shown as color-coded numerical values and the CAP256 mAbs are arranged with respect to their binding sensitivity to α2,6 desialylated CRF250 trimer. The four amino acids position in the CDRH2 (with respect to germline positions D53, K57, Y59 and K64) are indicated for each Ab. B. Percent (%) change in the octet binding responses are shown as heatmap for the desialylated trimer form and is calculated as (Ab binding response to the α2,6 desialylated CRF250 trimer/binding response to the CRF250 WT trimer). C. Phylogenetic tree showing the development of the CAP256 bnAb lineage from the VH3-30*18 germline gene. The branches in the tree are color-coded and demonstrate the evolution of amino acid residues at the above-mentioned four positions in CDRH2. The germline residues, DKYK and the most evolved residues DKHW in glycan-reactive Abs are indicated in green and red respectively. See also Figure S2C
