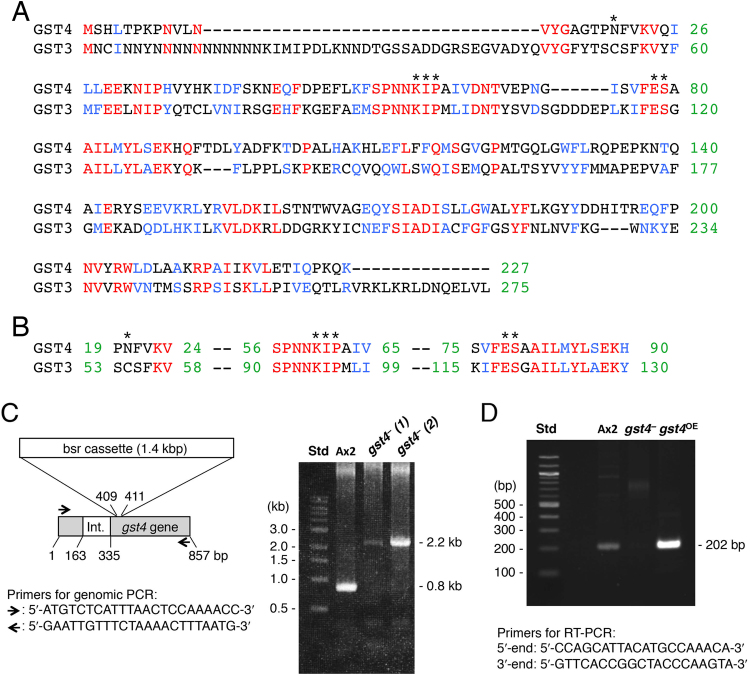
Fig. 3.
(A) Deduced amino acid sequences of GST3 and GST4. GST3 and GST4 were aligned by using the ClustalW program. Identical (red) and similar (blue) amino acid residues are indicated. The identity between the two GSTs was calculated to be 29.5%, their similarity was 55.9%, and the E-value was 2e−28 in BlastP. (B) Sequence alignment around the predicted GSH binding site. Asterisks mark the key conserved residues of the predicted GSH binding site [23]. (C) Generation of the gst4− mutant. gst4– cells were generated by using the indicated gene-targeting construct. Ax2 cells and 2 clones of the gst4– mutant underwent genomic PCR amplification. The PCR products were separated on an agarose gel and visualized by ethidium bromide staining. One of the clones, gst4–(2), was used for the present study. Int., intron; Std, molecular size standard. (D) Generation of the gst4OE mutant. The gst4OE mutant was generated as described in the Materials and Methods section, and gst4 mRNA expression in Ax2 cells and the gst4 mutants was assessed by semi-quantitative RT-PCR amplification with the indicated primers.