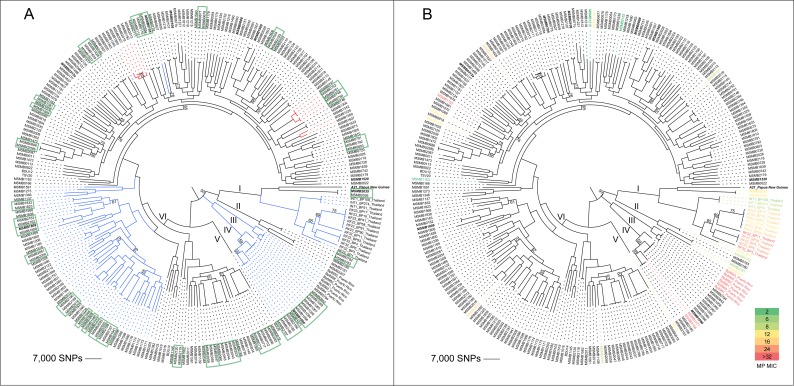
Fig 1. Phylogenomic analysis of 264 Burkholderia ubonensis genomes.
A midpoint-rooted maximum parsimony phylogeny was constructed using 589,433 biallelic core-genome SNPs. A) Strains lacking the B. pseudomallei multilocus sequence typing (MLST) gene narK are labelled with blue branches, and those lacking pC3 (previously known as chromosome III) are in bold italics. Highly related strains retrieved from single environmental samples are outlined by green boxes. Red branches indicate instances where isolates could be differentiated by the B. pseudomallei MLST scheme, but not the Bcc scheme. B) Heatmap of the meropenem minimum inhibitory concentration values for 40 tested B. ubonensis isolates. In both trees, the six distinct clades (I, II, III, IV, V and VI) are labelled. Consistency index = 0.25. Bootstrap values below 80% are labelled on their corresponding branches.