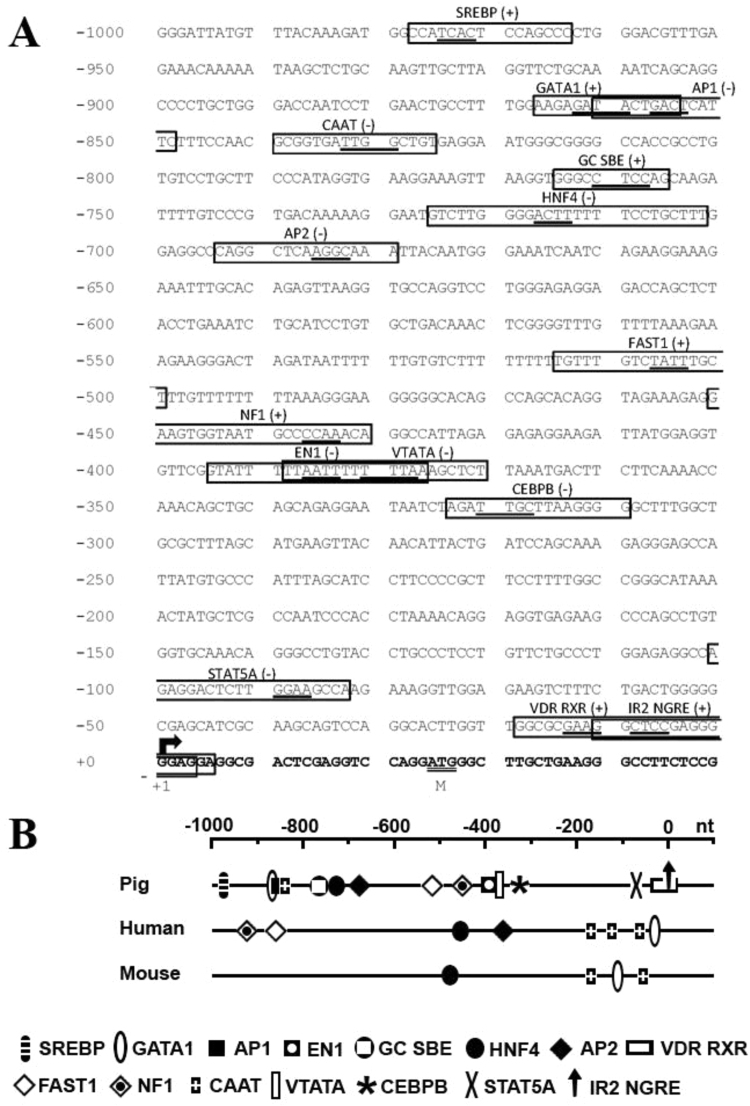
Fig. 8.
Location of putative transcription factor binding motifs in the pig SLC13A4 5’-flanking region. (A) Nucleotide sequence of the predicted SLC13A4 5’-flanking region (from −1000 to +50) is shown. Position +1 (arrow) denotes the putative transcription initiation site. The translation initiation ATG codon is double underlined. Boldface letters indicate the 5’-region of exon 1. A vertebrate conserved TATA box, CAAT-box, Vitamin-D binding site (VDR RXR) and GC- (SBE binding site) as well as putative transcription factor binding motifs are boxed, with core sequences underlined. Potential binding motifs were identified using MatInspector [23] with parameters of core >0.9 and matrix >0.8 similarities. SREBP, sterol regulatory element binding protein; GATA1, GATA binding factor 1; AP-1, associated protein 1; HNF4, hepatic nuclear factor 4; AP-2, associated protein 2; FAST1, FAST1 SMAD interacting protein; NF1, nuclear factor 1; EN1, homeobox protein engrailed; CEBPB, CCAAT/ enhancer binding protein beta; and STAT5A, signal transducer and activator of transcription factor 5. (B) Relative locations of each transcription-factor binding site were compared to those previously reported in the 5’-flanking region of human SLC13A4 and mouse Slc13a4[16], [26].