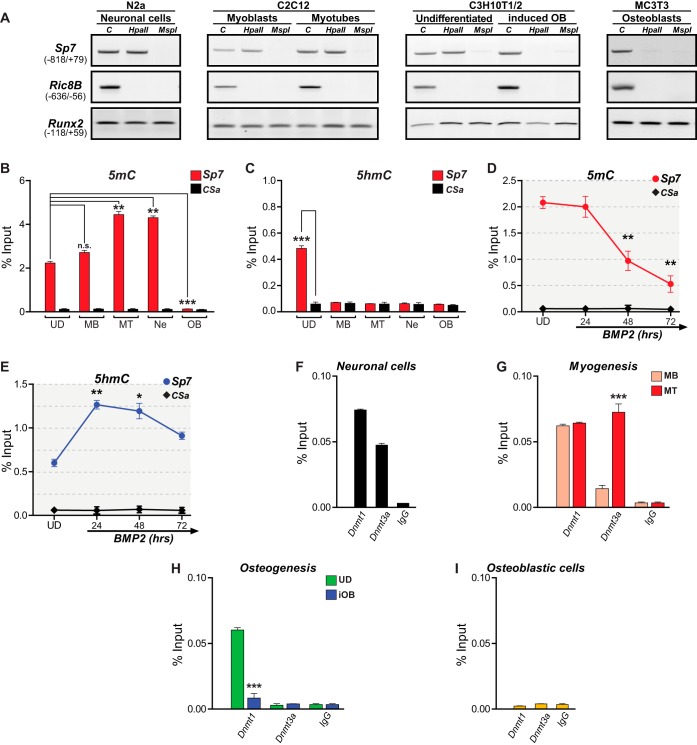
FIG 2.
Sp7 gene expression is directly associated with the Sp7 promoter DNA methylation/hydroxymethylation status. (A, B, and D) DNA methylation (5mC) at the Sp7 gene promoter in samples obtained from the indicated cell types was assessed by evaluating susceptibility to cleavage by the restriction endonucleases HpaII (sensitive to 5mC) and MspII (insensitive to 5mC) (A) as well as by MedIP analysis (B and D). (C and E) DNA hydroxymethylation (5hmC) was also determined by MedIP. In panel A, Ric-8B is a nonmethylated CpG-rich promoter sequence, and Runx2 is a low-CpG-containing promoter sequence that does not contain MspI/HpaII cleavage sites. In all MedIP analyses, CSa is a well-established unmethylated DNA sequence used as a control; MedIP values are expressed as a percentage of the input. (F to I) Enrichment of Dnmt1 and Dnmt3a at the Sp7 gene promoter determined by ChIP, using antibodies and chromatin samples isolated from Ne (F), MB and MT (G), UD and iOB (H), and OB (I) cells. ChIP values are expressed as a percentage of the input. See the legend to Fig. 1 for an explanation of abbreviations and symbols. Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., not significant (as determined by Student's t test).