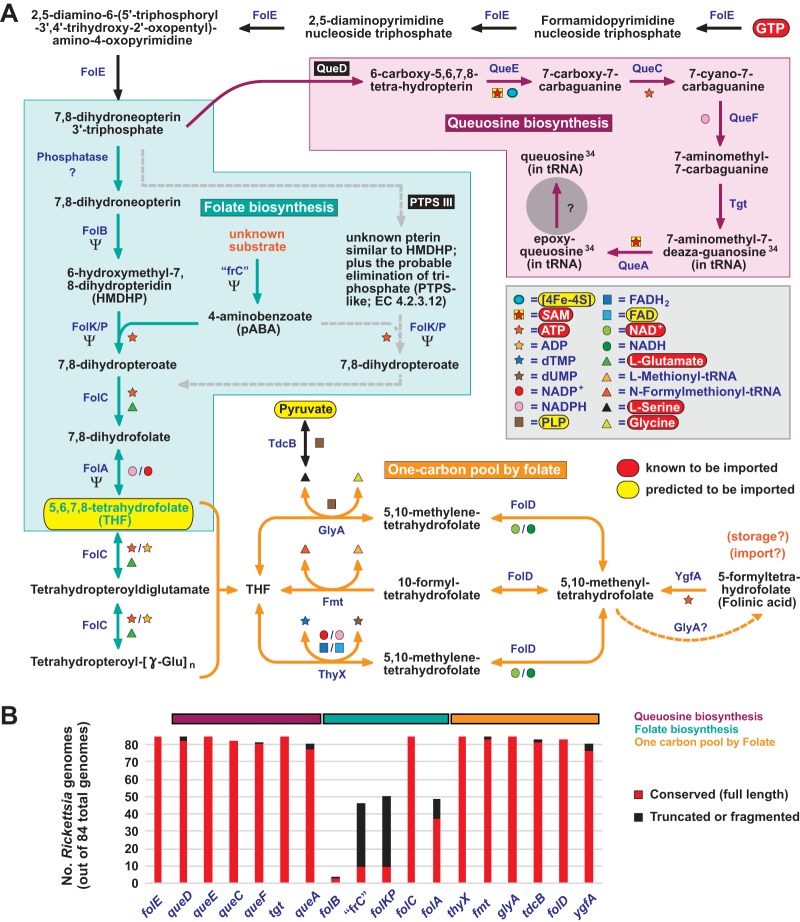
FIG 4 .
Rickettsia species lack the capability for de novo folate biosynthesis. (A) Rickettsiae use GTP cyclohydrolase I (FolE) to convert host-acquired GTP (red) to DHN-P3, a precursor of both queuosine (purple) and folate (aquamarine) biosynthesis. The classical THF synthesis pathway (aquamarine arrows), wherein DHN-P3 is dephosphorylated and subsequently converted to HMDHP by dihydropteridin aldolase (FolB), is disintegrating from Rickettsia genomes (Ψ denotes pseudogenization in over 50% of genomes). In the FolB bypass proposed by Hunter et al. (89) (gray dashed arrows), DHN-P3 is directly converted to HMDHP or a structurally similar molecule via PTPS-III (black box). Our analysis instead suggests that this enzyme is QueD (black box), which performs the first committed step in queuosine biosynthesis (see the text for further details). The one-carbon pool by folate (orange arrows) illustrates the role of host-acquired THF and several intermediates in the essential one-carbon transfer reactions that yield pyrimidine deoxynucleoside triphosphates, N-formylmethionyl-tRNA, and Ser/Gly. “frC,” fol_rel_CADD domain-containing protein (TIGR04305). The gray circle represents a hole in the pathway for queuosine synthesis (see Fig. S6). (B) Conservation of 18 genes involved in queuosine and THF biosynthesis and reactions within the one-carbon pool by folate. The complete distributions of these genes in 84 Rickettsia genomes reveals that no single Rickettsia species is capable of de novo folate biosynthesis, while the queuosine biosynthesis and one-carbon pool by folate pathways are highly conserved (see Fig. S5D).