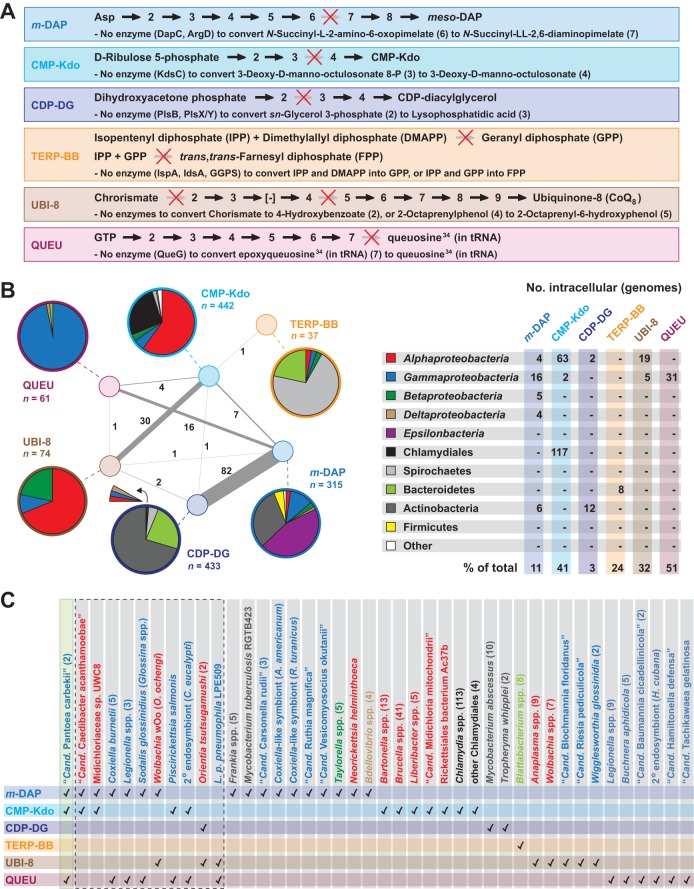
FIG 5 .
Comparative analysis of six biosynthetic pathways containing holes. The reconstructed Rickettsia metabolic network revealed holes in six biosynthetic pathways, DAP (m-DAP), CMP-Kdo, CDP-diacylglycerol (CDP-DG), terpenoid backbones (TERP-BB), ubiquinone-8 (UBI-8), and queuosine (QUEU). The distribution of these pathways across other prokaryotic genomes was determined via comparative metabolic pathway analyses. (A) Illustration and description of each hole-containing biosynthetic pathway. A red X indicates the missing enzyme(s) within each pathway, compared to well-characterized biosynthetic pathways for other prokaryotes. (B) Distribution of Rickettsia-like biosynthetic pathways across other prokaryotic genomes. The pie charts at the left indicate the taxonomic breakdown of genomes per biosynthetic pathway, with connections between pathways illustrating the number of genomes containing multiple Rickettsia-like pathways. Note that Rickettsia genomes were excluded from these analyses. At the right, the taxonomic color scheme is shown, with the number of genomes from intracellular species provided. (C) Intracellular species containing one or more Rickettsia-like biosynthetic pathways. The green box depicts the only genome found to contain three Rickettsia-like pathways, that of “Ca. Pantoea carbekii”) (117), which is an extracellular primary symbiont of the brown marmorated stink bug, where it is found in the gastric cecal lumina (see the text for further details). The dashed box indicates genomes containing two Rickettsia-like pathways. Taxa are colored in accordance with the color scheme in panel B.