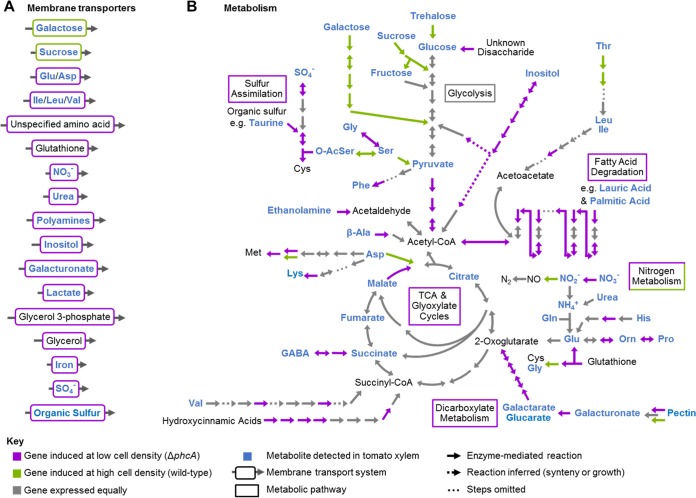
FIG 3 .
Differentially expressed genes that could enable R. solanacearum to use nutrients available in xylem sap. (A) Differentially regulated transporters. See Table S1B for specific gene expression levels. (B) Selected R. solanacearum metabolic pathways integrated with transcriptomic results. Metabolic pathways are based on KEGG analysis of the R. solanacearum GMI1000 genome. Compounds in blue are present in xylem sap (see references in Table S3). Purple arrows and boxes indicate reaction steps corresponding to enzymes whose expression was upregulated at low cell density (in the ΔphcA mutant); green arrows and boxes indicate enzymes whose expression was downregulated at low cell density (in the ΔphcA mutant).