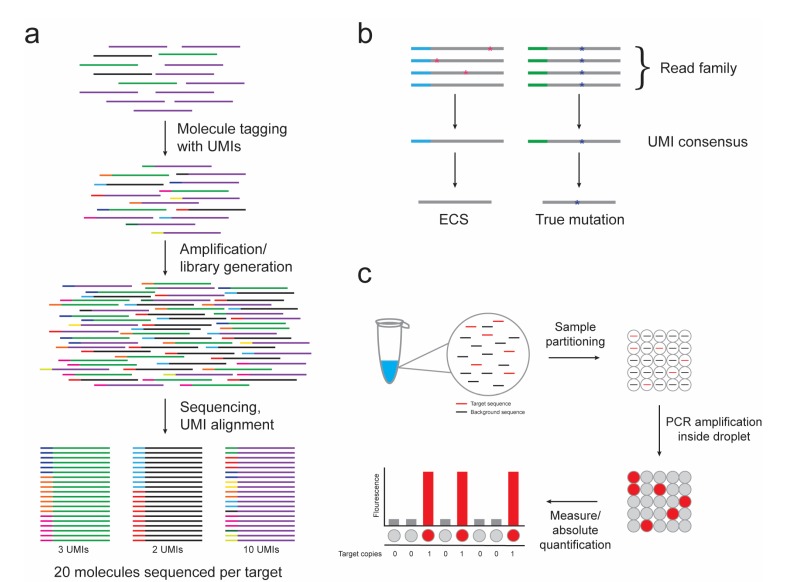
Figure 1.
The use of unique molecular indexes (UMIs) in next-generation sequencing (NGS) libraries and droplet digital PCR (ddPCR) provide technical advances for measurable residual disease (MRD) detection. (a) Molecules consisting of three different nucleic acid targets (purple, green, and black lines) are individually tagged with UMIs. The UMI tags are maintained throughout amplification and library generation and are used to count the number of original target molecules present in the sample despite PCR amplification bias. (b) Alignment of read families (grey line) sorted by UMIs (blue or green line) allows for the discrimination of true mutations (blue asterisk) from sporadic errors introduced during PCR or sequencing procedures (pink asterisk). (c) A single PCR reaction is partitioned by ddPCR into thousands to millions of water–oil droplets for which no more than a single target sequence (red line) is present per droplet. PCR amplification is performed within the droplets and the fluorescent signal is measured as either positive or negative, allowing for absolute target copy number quantification.