Fig. 4.
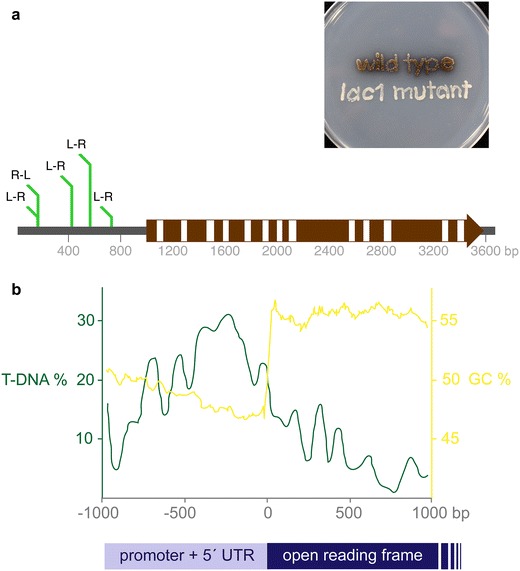
Two examples of T-DNA insertion bias into the 5′ non-coding (or flanking) regions of the genes in Cryptococcus neoformans (a basidiomycete yeast and human pathogen) and Pyricularia oryzae (a filamentous ascomycete and plant pathogen). a Example of a single gene in C. neoformans targeted by T-DNA insertion on five occasions [157, 159, 160] causing loss of pigmentation. A schematic drawing of the LAC1 gene represented by an arrow shows coding regions as brown boxes, introns as white boxes and upstream and downstream non-coding sequences as a grey line. T-DNA insertions from five independent transformation events each causing loss of pigmentation all lie within the upstream region, with none to date in the coding region. R and L refer to the left and right border and the relative positions of these when the T-DNA inserted; note that the same site is targeted by two independent insertion events in opposite orientation. The plate shows C. neoformans wild type and one of the T-DNA insertional mutants growing on medium containing the substrate for laccase, l-DOPA. b Example of insertional bias of T-DNA into the genome of P. oryzae transformants. Results and figure are modified from Ref. [40]. An analysis of the distribution of 799 insertion sites mapped into 50 bp windows illustrates a twofold higher insertion frequency in promoter or untranslated regions compared to coding regions. This is even more striking in that such regions have a higher proportion of AT nucleotides while in P. oryzae T-DNA insertions are preferentially into higher GC content DNA
