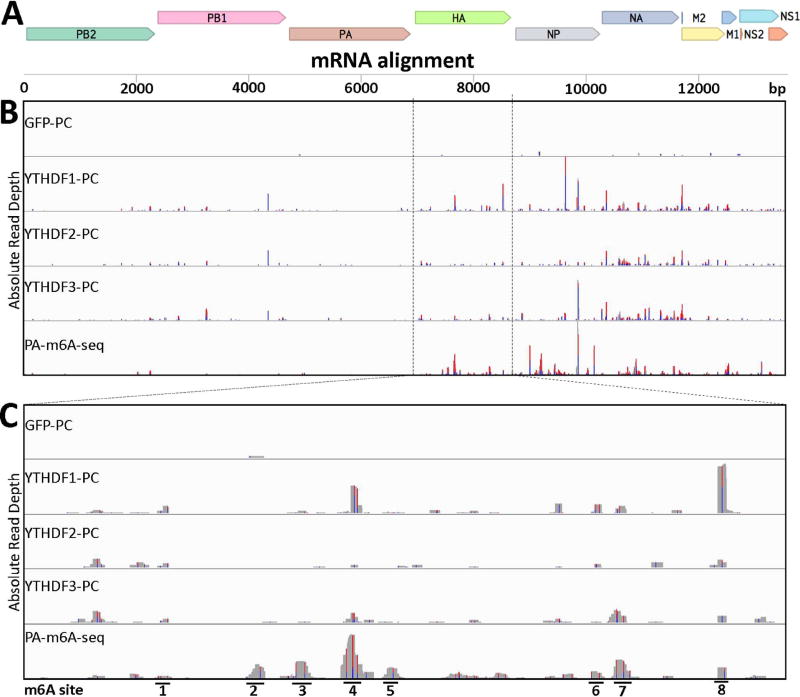
Figure 3. Identification of m6A sites on IAV-PR8 plus sense mRNA.
PAR-CLIP and PA-m6A-seq were performed using 293T cell lines 24 hpi with IAV-PR8 at an MOI of 10 (A) Concatenated map of the IAV-PR8 transcriptome that reads were aligned to. (B) Complete transcriptome coverage tracks are shown for PAR-CLIP performed on Flag-GFP, Flag-YTHDF1, Flag-YTHDF2 and Flag-YTHDF3 expressing 293T cells, while PA-m6A-seq was performed using wild type 293T cells. The PA-m6A-seq lane has a Y axis of 0–500 reads, and all others are depicted with Y axes of 0–200 reads. (C) An expanded view of the HA segment of IAV-PR8, with 8 prominent m6A sites numbered. PA-m6A-seq has a Y axis of 0–250 reads, and all others are depicted with Y axes of 0–100 reads. Reverse transcription of crosslinked 4SU residues results in characteristic T>C mutations and the level of T>C conversion at specific residues is indicated by red/blue bars.