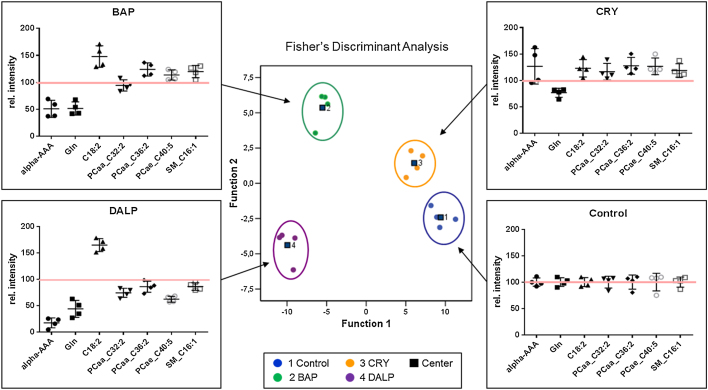
Fig. 4.
Fisher’s discriminant analysis of targeted metabolomics data obtained in HaCaT WT cells. Cells were treated with BAP (1 μM), CRY (1 μM) or DALP (0.01 μM) for 48 h in 4 replicates. Control samples contain 0.1% DMSO only. Metabolites of total cell extracts were analyzed using the p180 Kit from BIOCRATES. In HaCaT WT cells 24 metabolites contributed to a group separation between the three different PAH exposure groups and the control. Cross-validation of the data revealed a re-grouping of 100% using the leave-one-out system. Scatter dot plots for each exposure group (BAP, CRY and DALP) show the metabolic patterns of selected metabolites picked from the group of 24 metabolites that contributed to group separation. The short black lines refer to the mean values ± SD. The long red lines refer to the relative mean values of untreated controls which were set to 100%.