Figure 1.
DENV-3 Intrahost Diversity Is Shaped by Selection Pressures
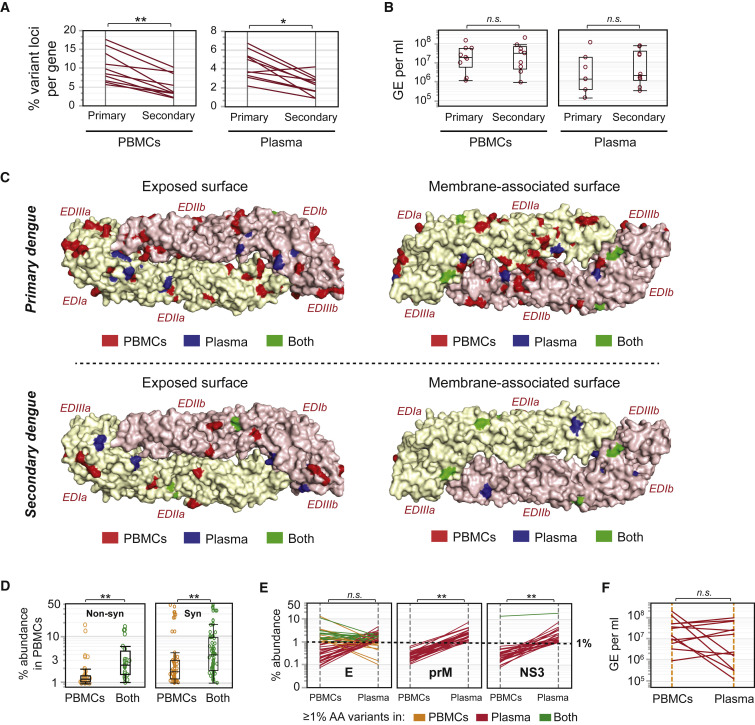
(A) Percent unique loci per protein with amino acid diversity, calculated as the percent of amino acids in a protein that show ≥1% variation, in PBMC (left panel) and plasma (right panel) samples from 53 individuals with 1° and 46 individuals with 2° dengue. ∗p ≤ 0.01, ∗∗p ≤ 0.001 (Wilcoxon signed-rank test). Each line represents one protein.
(B) Comparisons of genome equivalents per milliliter (GE/mL) of extracted RNA in 53 1° and 46 2° cases from PBMC (left panel) and plasma (right panel) samples, median ± SD, boxes represent 25th and 75th percentiles and whiskers are 10th and 90th percentiles. n.s., non-significant (Wilcoxon test for unpaired samples).
(C) All loci with ≥1% variation at the amino acid level, superimposed on the exposed surface (left panel) or the membrane-associated surface (right panel) of the DENV-3 E protein homodimer (PDB: 1UZG). Amino acids are colored according to whether they exhibit diversity in PBMC samples only (PBMCs, red), in plasma samples only (Plasma, blue), or in both PBMCs and plasma (Both, green). Data are shown separately for 53 1° (top panel) and 46 2° (bottom panel) dengue cases. EDIa/b, EDIIa/b, EDIIIa/b represent E domains I, II, and III from monomer chains a and b, respectively.
(D) Percent amino acid variants for loci that exhibit diversity in PBMCs only (PBMCs), compared with loci that exhibit diversity in both PBMCs and plasma (Both), median ± SD, boxes represent 25th and 75th percentiles and whiskers are 10th and 90th percentiles. In the left panel, the y axis represents the percent abundance of non-synonymous variants in PBMCs. In the right panel, the y axis represents the percent abundance of synonymous variants in PBMCs. ∗∗p ≤ 0.0001 (Wilcoxon test), n = 22 paired PBMC and plasma samples.
(E) Protein-specific comparisons of percent amino acid variants in 22 paired PBMC and plasma samples, colored by whether individual loci display ≥1% amino acid diversity in PBMC samples only (yellow), in plasma samples only (red), or in both PBMC and plasma samples (green). Data are shown for the two proteins (prM/M and NS3) that exhibit significant differences in % amino acid (AA) variants between PBMCs and plasma. All other proteins (including E, which is shown as an example) exhibit no significant differences in % AA variants between PBMCs and plasma. ∗∗p ≤ 0.0001, n.s., non-significant (Wilcoxon signed-rank test).
(F) Comparisons of genome equivalents per milliliter (GE/mL) of extracted RNA in 12 paired PBMC and plasma samples. n.s., non-significant (Wilcoxon signed-rank test for paired samples).