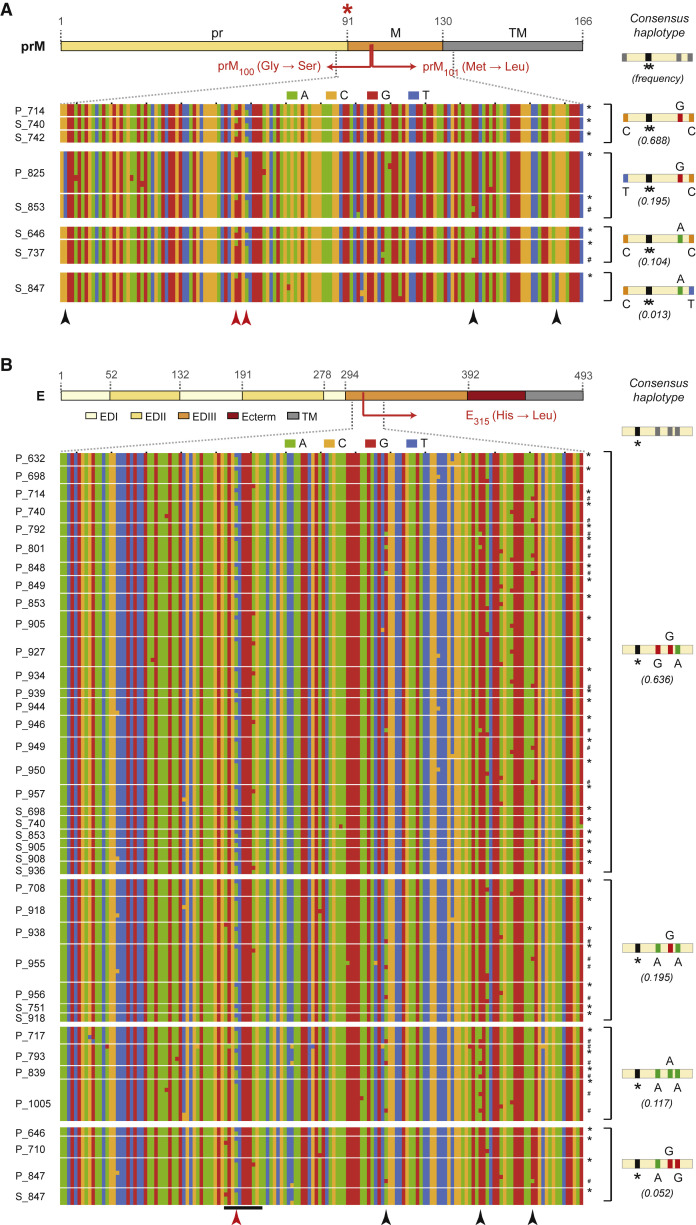
Figure 3.
Sequence Diversity in Reconstructed Haplotypes Spanning the Intrahost Diversity Hotspots in prM/M and E
Color-coded nucleotide diversity in regions flanking intrahost diversity hotspots in (A) prM/M and (B) E; each row represents a unique reconstructed haplotype with the hotspot variant in 8 PBMC/plasma samples for the prM/M hotspot and 39 PBMC/plasma samples for the E hotspot (samples delineated by thin white lines). Consensus haplotypes and their observed frequencies across all Nicaraguan samples are shown on the left, separated by thick white lines. Haplotypes with the prM100,101 and E315 variants are denoted with asterisks (∗), WT variant haplotypes distinct from the consensus haplotype are marked with #, and underlined loci in B represent the AB-loop region containing the E hotspot. Red asterisk, furin cleavage site. Red arrowheads, prM and E hotspot variants. Black arrowheads, other variants present at >1% abundance.