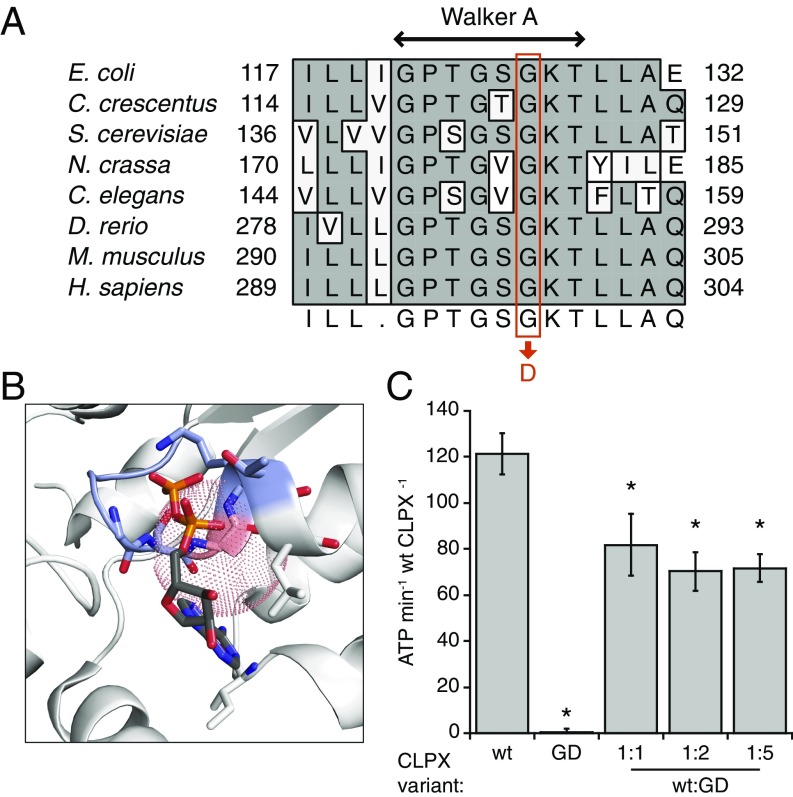
Fig. 2.
The CLPXGD mutation compromises the ATPase active site structure and activity. (A) The CLPXGD mutation occurs in the highly conserved Walker A ATPase motif. (B) Rendering of a nucleotide-binding pocket of ClpX (light gray) with the Walker A motif (blue-gray) and the residue corresponding to H. sapiens CLPX Gly298 (salmon). Coordinates are from a structure of E. coli ClpX6 bound to ADP (PDB ID code 3HWS) (38). The dotted sphere represents the maximum extension of an aspartate side chain from this position (3.72 Å). ADP atoms are colored using the standard scheme, and side chain residues that form hydrogen bonds with ADP are also shown. (C) ATPase activity of CLPX, CLPXGD, and mixtures thereof. ATPase activity of 0.15 µM (hexamer concentration) CLPX (WT), 0.15 µM CLPXGD (GD), or 0.15 µM CLPX plus 0.15, 0.3, or 0.75 µM CLPXGD (WT + GD) is displayed. Activity is calculated relative to the concentration of WT CLPX in each condition (ATP per minute per WT CLPX6). Mean values ± SD are plotted; n ≥ 3 biological replicates. *Reduced activity, P ≤ 0.005, Student’s t test.