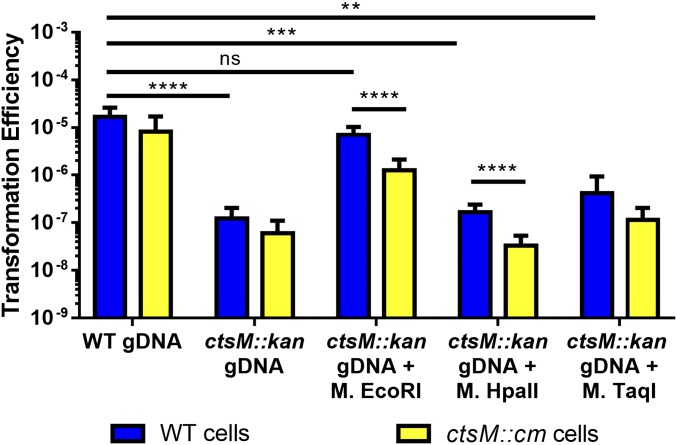
Fig. 5.
In vitro methylation at the CtsM site RAATTY, but not at other sites, significantly enhances gDNA substrate capability. DNA was purified from either DRH153 (kanr) or ctsM::kan cells. ctsM::kan DNA was then either mock-methylated or in vitro methylated with EcoRI MTase, TaqI MTase, or HpaII MTase. EcoRI MTase methylates GAm6ATTC, TaqI methylates TCG m6A, and HpaII methylates C m6CGG. WT cells (blue) or ctsM::cm cells (yellow) were transformed with the methylated DNA. Transformation efficiency was calculated as described previously. Data represent three biological replicates, each of which contained three technical replicates. Error bars indicate SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Kruskal–Wallis test with Dunn’s multiple-comparisons test. ns, not significant.