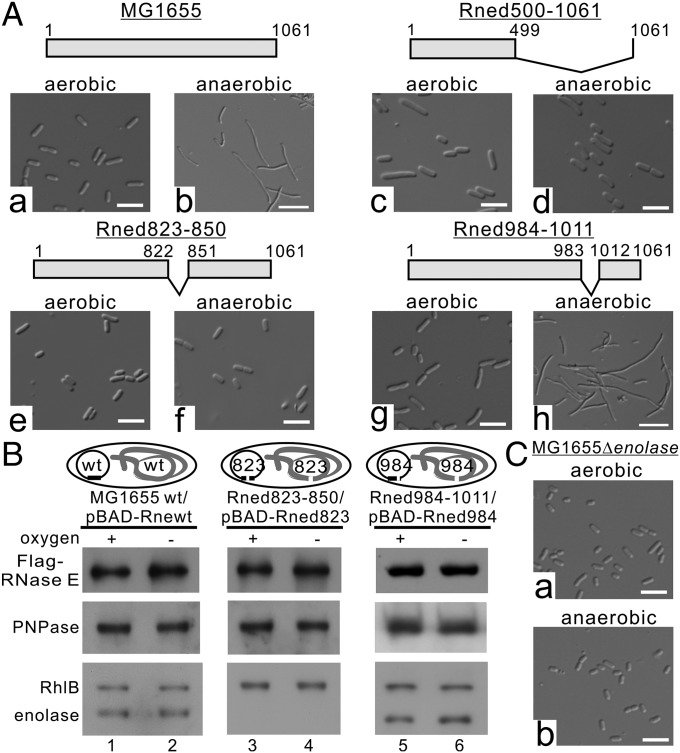
Fig. 2.
Cell filamentation under anaerobic conditions requires enolase binding to RNA degradosomes. (A) Cell morphology of E. coli MG1655 (a and b) and its derivative strains (c–h) under aerobic and anaerobic conditions. Schematic representations of RNase E and its endogenous (untagged) and plasmid-encoded (Flag-tagged) variants are presented above the respective images. (Scale bars, 5 µm.) (B) The degradosomes isolated from E. coli MG1655 and its derivatives, Rned823–500 and Rne984–1011. Protein aliquots containing equal amounts of Flag-RNase E and its variants were analyzed by SDS/PAGE and immunoblotted with the indicated antibodies. Schematic representations of RNase E and its endogenous (untagged) and plasmid-encoded (Flag-tagged) variants are presented above the respective images. (C) Cell morphology of the E. coli MG1655Δenolase mutant with endogenously deleted enolase under aerobic and anaerobic conditions. (Scale bars, 5 µm.)