Significance
We present an epigenetic switch in the central control of reproduction as a truncated TET1, expressed in proliferating gonadotrope-precursor cells, which inhibits Lhb expression and so must be repressed for reproductive development. Expression of this TET1 isoform is regulated by cis-elements mediating effects of gonadal steroids and PKA, and also a potentially methylated distal enhancer. As this isoform appears more common than the canonical TET1 in other differentiated tissues, our study has broader functional implications outside of the reproductive axis. Furthermore, our findings support the idea that distinct genomic regions are used at different developmental stages or in different tissues, and that a particular sequence can be part of the primary transcript in some tissues or an enhancer RNA in others.
Keywords: Tet1, Tet2, gonadotrope, enhancer, luteinizing hormone
Abstract
The TET enzymes catalyze conversion of 5-methyl cytosine (5mC) to 5-hydroxymethyl cytosine (5hmC) and play important roles during development. TET1 has been particularly well-studied in pluripotent stem cells, but Tet1-KO mice are viable, and the most marked defect is abnormal ovarian follicle development, resulting in impaired fertility. We hypothesized that TET1 might play a role in the central control of reproduction by regulating expression of the gonadotropin hormones, which are responsible for follicle development and maturation and ovarian function. We find that all three TET enzymes are expressed in gonadotrope-precursor cells, but Tet1 mRNA levels decrease markedly with completion of cell differentiation, corresponding with an increase in expression of the luteinizing hormone gene, Lhb. We demonstrate that poorly differentiated gonadotropes express a TET1 isoform lacking the N-terminal CXXC-domain, which represses Lhb gene expression directly and does not catalyze 5hmC at the gene promoter. We show that this isoform is also expressed in other differentiated tissues, and that it is regulated by an alternative promoter whose activity is repressed by the liganded estrogen and androgen receptors, and by the hypothalamic gonadotropin-releasing hormone through activation of PKA. Its expression is also regulated by DNA methylation, including at an upstream enhancer that is protected by TET2, to allow Tet1 expression. The down-regulation of TET1 relieves its repression of the methylated Lhb gene promoter, which is then hydroxymethylated and activated by TET2 for full reproductive competence.
TET enzymes catalyze the conversion of 5-methyl cytosine (5mC) to 5-hydroxymethyl cytosine (5hmC), which blocks some of the 5mC repressive effects while also catalyzing additional modifications of 5hmC to bases that are quickly removed, thus comprising a pathway to active demethylation (1–4). However, the TET proteins are found enriched at CpG-rich gene promoters and 5hmC is readily detected in many cell types, particularly at active or poised regulatory elements, suggesting a facilitating role in transcriptional activation (5–7).
Although the three TET proteins harbor the same catalytic activity, they are expressed in distinct developmental and tissue-specific patterns (8). TET1 is at its highest levels in ES cells (ESCs) and is down-regulated during differentiation. TET2 is also expressed in ESCs, and transcription of both genes is regulated by the pluripotency factors (9, 10). Despite this, both TET1 and TET2 are also expressed through largely unknown mechanisms in differentiated tissues, although mice lacking TET1 or TET2 are viable and, asides from the TET2 effects on hematopoiesis, their definitive roles are mostly not clear (11–13). In fact, the most overt phenotype of TET1-KO mice was a defect in ovarian development: their ovaries were small and had fewer mature follicles and reduced fertility, even though embryonic germ-cell development appeared normal (11, 12), pointing to a unique role for TET1 in the regulation of follicular development.
Ovarian growth and activity are regulated by the gonadotropins luteinizing hormone and follicle-stimulating hormone. These hormones are expressed in the pituitary gonadotropes during embryonic development, and are up-regulated during a neonatal period of proliferation, but then become quiescent until puberty, when they are reactivated by the hypothalamic gonadotropin-releasing hormone (GnRH). We have shown previously that specific chromatin modifications are involved in determining the expression of these genes in a tissue-specific or hormonally induced context (14–19). However, the Lhb gene promoter is particularly rich in CpGs, prompting us to consider that it might be regulated by DNA methylation, and the effect of TET1 KO on ovarian development and function opened the possibility that this might be modified by TET1.
Our study revealed that an N-terminal truncated TET1 is expressed in poorly differentiated proliferating gonadotropes, which appears as the more common isoform in other differentiated tissues; it is regulated by an alternative proximal promoter and repressed by estradiol (E2) and dihydrotestosterone (DHT), and also by GnRH via activation of PKA. Its expression can also be regulated by methylation, including at an upstream enhancer that is protected by TET2. TET1 down-regulation in the gonadotrope precursor cells relieves repression of the Lhb gene promoter, which is then hydroxymethylated by TET2 to allow Lhb expression and reproductive competence.
Results
Tet1 Negatively Correlates with Lhb Expression and Is Down-Regulated by GnRH and Gonadal Steroids.
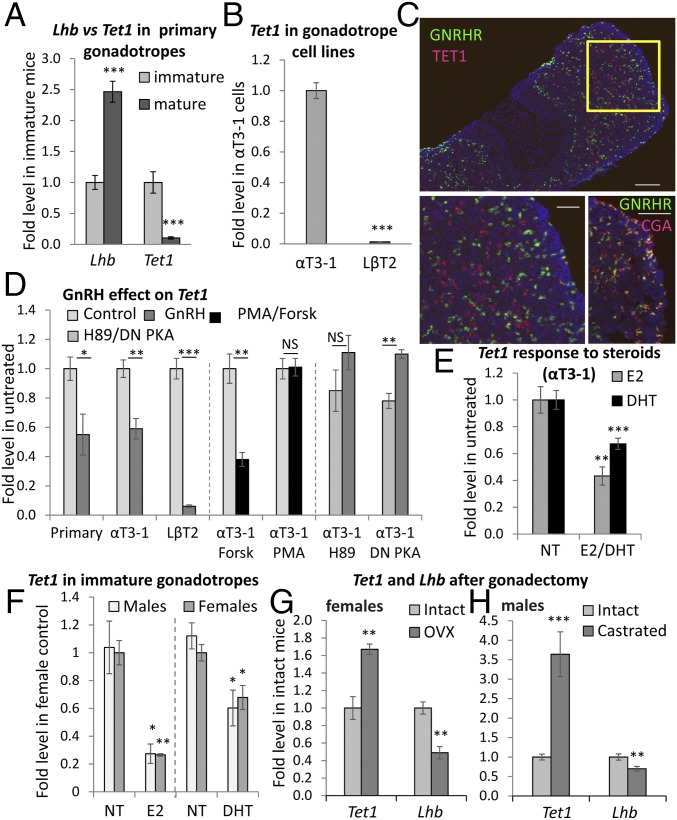
We detected all three Tet mRNAs in primary murine gonadotrope cells. However, the levels of Tet1 were considerably higher in cells from immature 6-d-old mice in which the gonadotrope population is expanding than in the gonadotropes of adult (8–14 wk) sexually mature mice, whereas the levels of Tet2 and Tet3 mRNAs were similar in both groups (Fig. S1A). Thus, there is a negative correlation of Tet1 with the mRNA levels of Lhb, which is expressed at a higher level in the adult mice (Fig. 1A). In gonadotrope cell lines, Tet1 levels were much lower in the more fully differentiated LβT2 cells in which Lhb is expressed abundantly than in the poorly differentiated αT3-1 cells in which Lhb is barely expressed (Fig. 1B) (17), whereas levels of Tet2 and Tet3 differed less (Fig. S1B). This negative correlation of Tet1 expression and gonadotrope maturation was confirmed in pituitary sections from adult mice, in which TET1 was notably lacking in the mature gonadotropes (Fig. 1C). Given that TET1 expression in the immature pituitary appears restricted to CGA-positive cells (Fig. S1C), and is found in the adult pituitary but not in the gonadotropes, these cells are very likely thyrotropes.
Fig. S1.
Tet1 expression negatively correlates with gonadotrope differentiation (Fig. 1). (A and B) qPCR analyses of relative levels of each of the TET enzyme mRNAs in (A) primary gonadotropes from immature (6 d) or mature (8–14 wk) mice or (B) partially differentiated (αT3-1) or fully differentiated (LβT2) gonadotrope cell lines; Tet mRNA levels were calculated by using standard curves of plasmid cDNA (n = 3–9). A t test was used to compare levels of the same gene between groups of mice (**P < 0.01 and ***P < 0.001; NS indicates P > 0.05). (C) TET1 (red) and CGA (green) in the pituitary of 5-d-old female mouse shown at lower magnification (Top Left) and subsequently the boxed region enlarged for each color channel separately or overlaid (Bottom Right). (Scale bars: lower magnification, 100 µm; higher magnification, 50 µm.) Notably, TET1 is seen exclusively in CGA-expressing cells at this stage, but is not found in all CGA+ cells. (D and E) Tet2, Tet3, and Fshb mRNA levels in the OVX (D) or castrated mice (E) in Fig. 1 G and H shown relative to levels in the intact mice; there are no significant differences between means of the two groups for any of these genes (P > 0.05). (F) Gonadotrope cells from intact females (freely cycling) and OVX mice were cultured with E2 for 48 h before qPCR analysis of Tet1 mRNA levels as before (n = 3; *P < 0.05; NS indicates P > 0.05).
Fig. 1.
Tet1 negatively correlates with Lhb expression and is down-regulated by GnRH and gonadal steroids. (A and B) qPCR analyses in (A) primary gonadotropes from immature (6 d) or mature (8–14 wk) mice or (B) αT3-1 and LβT2 cells; mRNA levels are relative to immature mice or αT3-1 cells (n = 3–9). A t test was used to compare levels of the same gene between cells (***P < 0.001). (C) TET1 (red) in the adult pituitary of GRIC-GFP mice; GnRHR+ cells appear green. (Scale bar: 100 µm). (Bottom Left) Enlargement of boxed region; (Bottom Right) CGA (red) in the same pituitary. (Scale bars: 50 µm.) (D) Tet1 levels in cell lines and primary cells from immature mice after exposure to GnRH in culture; αT3-1 cells were also exposed to forskolin or PMA; alternatively treated with H89 or transfected with dominant-negative (DN) PKA with and without GnRH exposure (n = 2–6; *P < 0.05 and ***P < 0.001; NS indicates P > 0.05; all means shown relative to untreated controls). (E) αT3-1 cells or (F) primary gonadotropes from immature mice were exposed to E2 or DHT; Tet1 mRNA levels are presented as in Fig. 1D (n = 4–6). (G and H) Tet1 and Lhb mRNA levels in gonadotropes 10 d after (G) ovariectomy (n = 4) or (H) castration (n = 8) shown relative to age-matched intact controls (Fig. S1).
Given that the levels of Tet1 gene expression shifted in accordance with a change in Lhb expression, we considered that regulatory hormones along the reproductive axis might play a role in determining its levels. Indeed, GnRH, the major activator of Lhb gene expression, reduced Tet1 mRNA levels in both cell lines and primary cells. As we were able to mimic this effect by incubation of the cells with forskolin but not phorbol 12-myristate 13-acetate (PMA), we considered that the GnRH effect is via activation of PKA, which was confirmed by pretreatment with H89 or transfection of a dominant-negative PKA, both of which completely abolished the GnRH repressive effect, and there was even a slight increase in Tet1 expression (Fig. 1D).
We also exposed the gonadotrope cell lines and primary cells from immature mice of both sexes to E2 or DHT (10 nM, 48 h). Both steroids repressed Tet1 expression, although E2 appeared more potent (Fig. 1 E and F). To further understand the relationship between TET1 and the reproductive axis, we ovariectomized (OVX) or castrated adult mice to remove the gonadal steroid feedback to assess the effect on Tet1 expression. As in the neonatal mice, the gonadotropes are proliferating in this state and include immature precursor cells (20). Tet1 mRNA levels were significantly increased in these cells, especially in the males, although those of Tet2 and Tet3 were not affected (Fig. 1 G and H and Fig. S1 D and E). In males and females, average levels of Lhb were reduced in this precursor cell population compared with the fully differentiated population in intact mice (Fig. 1 G and H), whereas the levels of Fshb were unaltered (Fig. S1 D and E). In cells from the OVX mice, E2 reduced the elevated Tet1 expression, whereas no effect was apparent in cells from intact adult females (Fig. S1F).
The Gonadotrope Precursor TET1 Isoform is N-Terminally Truncated, Found in Other Differentiated Tissues, and Uses an Alternate Promoter That Binds the Liganded ESR1 and AR.
In PCR analyses of the Tet1 mRNA in both cell lines and primary gonadotropes (from immature mice), we could barely detect the first coding exon 1 of the canonical Tet1, but an alternative exon, which we termed exon 1.5, was amplified (Fig. 2 A and B). Analysis revealed this same pattern of exon expression in olfactory bulb and mammary glands, whereas, in the cerebellum, cerebral cortex, liver, and heart, both exons were detected but at differing ratios, and in the placenta, as in ESCs, exon 1 was expressed at a much higher level (Fig. S2 A and B). RACE confirmed that the functional Tet1 transcriptional start site (TSS) in the gonadotropes is indeed located 181 bp upstream of this exon 1.5 (Fig. S2 C and D).
Fig. 2.
The gonadotrope precursor TET1 isoform is N-terminally truncated, found in other differentiated tissues, and uses an alternate promoter that binds liganded ESR1 and AR. (A) RT-PCR analyses for the Tet1 canonical exon 1, the alternative exon “1.5,” and the canonical exon 3 in mouse ESCs, αT3-1 and LβT2 cell lines, and gDNA, with Gapdh control. (B) qPCR of αT3-1 cells and primary gonadotropes from immature mice using the same primer sets and quantified by using a standard curve of gDNA, shown relative to levels of exon 3 (n = 5). (C–G) ChIP analysis for (C) H3K4me3 and (D) H3K27ac in αT3-1(Left) and LβT2 (Right) cells for the region upstream of the canonical exon 1 (−14,665 to −14,572 bp) and exon 1.5 (−53 to +68) or in (E) RNAPII S5p, (F) ESR1, and (G) AR in αT3-1 cells (exposed for 2 h to E2 or DHT as noted). IP levels are relative to input (n = 3; Figs. S2 and S3).
Fig. S2.
The gonadotrope Tet1 isoform lacks exon 1, which encodes the CXXC domain (Fig. 2). (A) RT-PCR of the mouse brain (C, cerebellum; CC, cerebral cortex; OB, olfactory bulb) or placenta (marked as “P”) or (B) other mouse tissues using the same primer sets. G, GAPDH; NTC, no template control. (C) RT-PCR was carried out on αT3-1 cells using sets of primers targeting the first three possible exons of Tet1 as shown. (D) 5′RACE was carried out on αT3-1 cell cDNA, with nested primers targeting exon 3 of Tet1. The amplification product of the second round of PCR was resolved by gel electrophoresis, and the ∼500-bp product was identified by sequencing. The actual sequence is shown, with the start of the published sequence underlined.
ChIP analysis for H3K4me3 and H3K27ac was carried out to determine whether this region of the Tet1 gene carries histone marks characteristic of active promoters. Both modifications are clearly enriched at this region, and at much higher levels in αT3-1 than in LβT2 cells, but were not seen in either cell line around the canonical first exon (Fig. 2 C and D). Cap analysis of gene expression (CAGE) data (FANTOM5) also indicates that this is the TSS for Tet1 in other tissues, whereas the Encyclopedia of DNA Elements (ENCODE) data shows dual peaks of H3K4me3 and H3K27ac at this location, as well as H3K4me3 at a region upstream of the canonical exon 1 in all tissues except the ESCs and placenta, in which only the upstream peak is apparent (Fig. S3). This clearly indicates that TET1 can be expressed as a short or long isoform in distinct tissues, but, in many, including the immature gonadotropes, the dominant form is the short TET1 that lacks the CXXC domain.
Fig. S3.
The gonadotrope-truncated Tet1 isoform arises from an alternative TSS and appears common in differentiated tissues (Figs. 2 and 4). The region upstream of Tet1 is shown in the University of California, Santa Cruz (UCSC), genome browser, with CAGE data for transcript initiation sites and the locations of (A) H3K4me3 and (B) H3K27ac in various tissues; the core of CpGI 1 is marked in green.
The functional promoter of this TSS was demonstrated finally by ChIP, in which RNAPII S5p was seen to bind (Fig. 2E), as did the liganded ESR1 and AR; ESR1 was enriched at −550 bp upstream and also around −1.4 kbp, whereas AR appeared enriched throughout this region (Fig. 2 F and G).
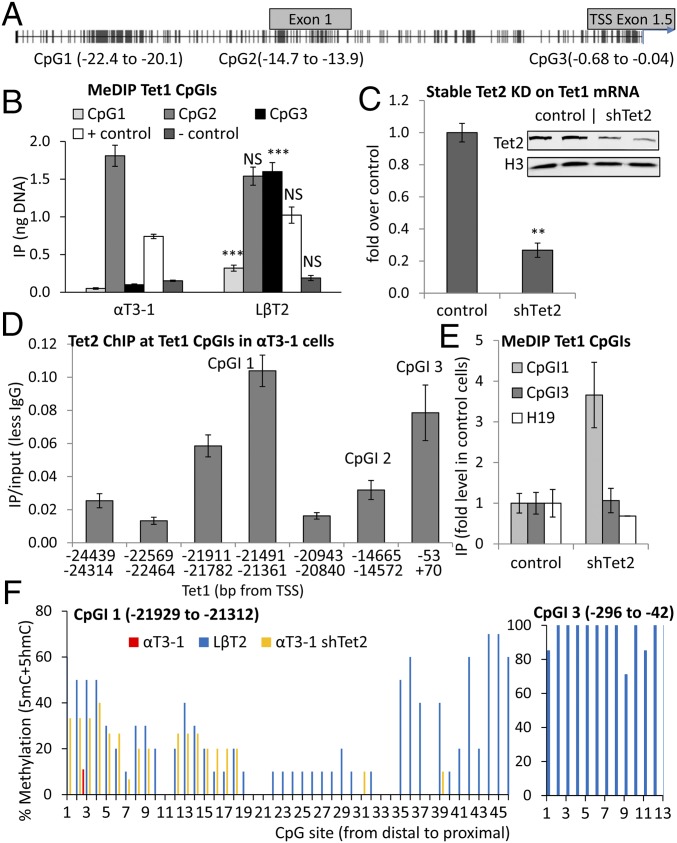
The Region Upstream of Tet1 TSS Can Be Methylated at Three Distinct CpGIs, but the Distal Part of the Most 5′CpGI Is Protected by TET2.
We postulated that the use of a different promoter by Tet1 in the gonadotrope precursor cells might be the result of DNA methylation. Three CpG islands (CpGIs) identified by Methyl Primer Express Software (Applied Biosystems) using default parameters were found: CpGI 1 at a distal site 22.4 kbp upstream of the functional Tet1 TSS, CpGI 2 just upstream of the canonical exon 1, and CpGI 3 immediately upstream of the functional TSS (Fig. 3A). Methylation at CpGI 1 and 3 was much higher in LβT2 than in αT3-1 cells, whereas CpGI 2 was highly methylated to a similar level in both cell types (Fig. 3B), indicating that Tet1 expression might well be coupled to methylation of each of these regions.
Fig. 3.
The region upstream of this Tet1 TSS can be methylated at three distinct CpGIs, but the distal part of the most 5′ CpGI is protected by TET2. (A) CpGIs upstream of Tet1 relative to the functional TSS. (B) Levels of 5mC DNA (relative to gDNA standard curve) at these CpGIs (n = 4–5). Statistical analysis (as in Fig. 1) compared each region between cell lines. (C) Tet1 mRNA levels after TET2 KD (shTet2) in αT3-1 cells; data analyzed and presented as before (n = 3–4). Western blot analysis shows the TET2 KD (n = 2). (D) ChIP of TET2 at the CpGs in αT3-1 cells, analyzed and presented as previously (n = 4–7). (E) MeDIP for the core of CpGI 1 (−21,491 to −21,361 bp) and CpGI 3 (−2 to −191 bp) from WT αT3-1 or shTet2 cells shown as fold in control cells, with H19 as unaffected positive control (n = 4–5). (F) BS analysis of Tet1CpGIs 1 and 3 in the two cell lines and for CpGI 1 in shTet2 cells; percentage of each CpG was methylated in 6–15 independent clones from each cell line.
TET proteins have been reported to protect CpGIs from DNA methylation (21), and stable TET2 knockdown (KD) reduced Tet1 expression by ∼80% (Fig. 3C), suggesting its possible role in regulating Tet1. ChIP revealed that, in the immature αT3-1 cells, TET2 is indeed enriched at the two regions, corresponding to CpGIs 1 and 3 (Fig. 3D). Furthermore, methylated DNA immunoprecipitation (MeDIP) analysis showed that the Tet1CpGI 1 is nearly fourfold more methylated in Tet2 KD than in control cells, although CpGI 3 was unaffected (Fig. 3E), clearly suggesting that TET2 plays a pivotal role at this distal CpGI 1.
To confirm these findings and clarify which regions of CpGIs 1 and 3 are methylated, we analyzed the core of CpGI 1 (−22 to −21.3 kbp) and CpGI 3 (−296 to −42 bp) in bisulfite (BS)-converted DNA from control αT3-1 and LβT2 cells, and also at the core of CpGI 1 in the DNA from TET2-KD cells. The methylation was much higher in LβT2 cells than in the control αT3-1 cells at both CpGIs (Fig. 3F). In the TET2 KD cells, there was a clear increase in the methylation at CpGI 1 compared with the control cells, but only in the most distal part (P < 0.001, Fisher’s exact test; Fig. 3F), suggesting that TET2 inhibits DNA methylation at this region, whereas the methylation at the proximal part of this CpGI, as well as the promoter (CpGI 3), is regulated via other mechanisms.
The Upstream CpGI Likely Comprises a Transcriptional Enhancer.
These findings in which methylation of CpGI 1 was associated with very low levels of Tet1 indicate that this distal region plays a role in regulating Tet1 expression, possibly acting as a transcriptional enhancer. We therefore performed ChIP analysis for H3K4me1, H3K4me3, and H3 on a single batch of αT3-1 cells. The distribution of the two H3K4 modifications clearly differed, with the core of the CpGI being enriched for H3K4me3 but not for H3K4me1 (Fig. 4A), whereas the region 5′ to the CpGI had higher levels of H3K4me1 and much lower levels of H3K4me3 (∼40-fold decrease). Thus, a distinct region at the 5′ end of the CpGI contains a particularly high ratio of H3K4me1/H3K4me3, indicating that its likely function as an enhancer (22). However, this region was not enriched for H3K27ac (Fig. 4B), similar to other tissues expressing primarily the short Tet1 isoform (Fig. S3B).
Fig. 4.
The upstream CpGI likely comprises a transcriptional enhancer. (A and B) ChIP analysis of CpGI 1 for (A) H3K4me1, H3K4me3, and H3 or (B) H3K27ac, H3, and IgG in αT3-1 cells, as in Fig. 2 (n = 2–4; Fig. S3). (C) Total αT3-1 RNA was reverse transcribed (RT) for qPCR of the regions marked. Controls lacked reverse transcription (normalized to gDNA standards; n = 3; Fig. S4). (D–G) Levels of eRNA were measured similarly in (D) αT3-1 and LβT2 cells; (E) WT and shTet2 αT3-1 cells; (F) LβT2 cells with or without GnRH treatment; and (G) primary gonadotrope cells (n = 3–6). (H and I) A 3C assay was carried out in Dpn2-digested DNA from αT3-1 cells, and chimeric fragments were detected by using nested forward primers (nos. 842 and 843 or nos. 844 and 845) targeting the functional TSS or upstream of the canonical exon 1, with various primers targeting the upstream region, as detailed in Fig. S4. (H) Amplicons were resolved by electrophoresis and identity confirmed by sequencing or (I) measured by qPCR using standard curves of the cloned chimeric fragments (n = 4–10); a t test was used to compare interaction of the pairs of regions.
Active enhancers often produce noncoding RNAs [i.e., enhancer RNAs (eRNAs)], which may play crucial roles in their activity (e.g., refs. 18, 23). The region just upstream of the 5′ end of the CpGI 1 was transcribed at a much higher level than the surrounding region (Fig. 4C and Fig. S4). Moreover, the level of this eRNA was higher in αT3-1 than in LβT2 cells, consistent with the levels of the Tet1 transcript (Fig. 4D), whereas, in TET2 KD cells, the eRNA levels, like those of the Tet1 mRNA, were reduced (Fig. 4E). In both cases, the lower Tet1 mRNA and eRNA levels corresponded with elevated levels of 5mC at the distal CpGI (Figs. 1B and 3B), suggesting that DNA methylation is involved in regulating expression of the eRNA. Despite the dramatic decrease in Tet1 expression following GnRH exposure in LβT2 cells (Fig. 1D), the eRNA levels did not change (Fig. 4F). Also in primary cells, eRNA levels were lower in gonadotropes of mature than immature mice (Fig. 4G), in accordance with the levels of the Tet1 transcript (Fig. 1A). These findings confirm that this region is likely an enhancer in which transcription of the eRNA is not determined directly by that of Tet1, but it may form part of a basal regulatory mechanism.
Fig. S4.
Characterization of the Tet1 distal transcriptional enhancer (Fig. 4). (Top) The region upstream of Tet1 is shown in the UCSC genome browser, with the core of CpGI 1 marked in green and the CAGE data for transcript initiation sites. The region transcribed to eRNA is shown together with the locations of various primers used for its amplification (primers marked in red were successful and those in black were not). (Bottom) The region upstream of the Tet1 functional TSS in the gonadotropes is marked, with vertical arrows signifying Dpn2 restriction sites and horizontal arrows the location of primers used in the 3C assay. The numbers in red refer to the primer identity. “Ex 1” refers to the canonical exon 1, and the CpGI is CpGI 1 at the region of the enhancer.
Finally, we performed chromatin conformation capture (3C) to assess whether this region interacts with the functional TSS. PCR was carried out on a Dpn2-digested and ligated 3C library using nested primers for semiquantitative analysis or individual primer sets for quantitative PCR (qPCR; Fig. S4). One set of primers targeted the functional TSS region (842 and 843; −221 bp and −191 bp) and the other targeted just upstream of exon 1 (844 and 845; −14,991 and −15,011 bp from the TSS). These were used with various upstream primers. Both methods showed interaction of the distal CpGI 1 region with the functional TSS, far more than with the region upstream of exon 1. This difference was further quantified by qPCR, normalized to the same cloned chimeric fragment, thus taking into account differences in the primer efficiency (Fig. 4 H and I).
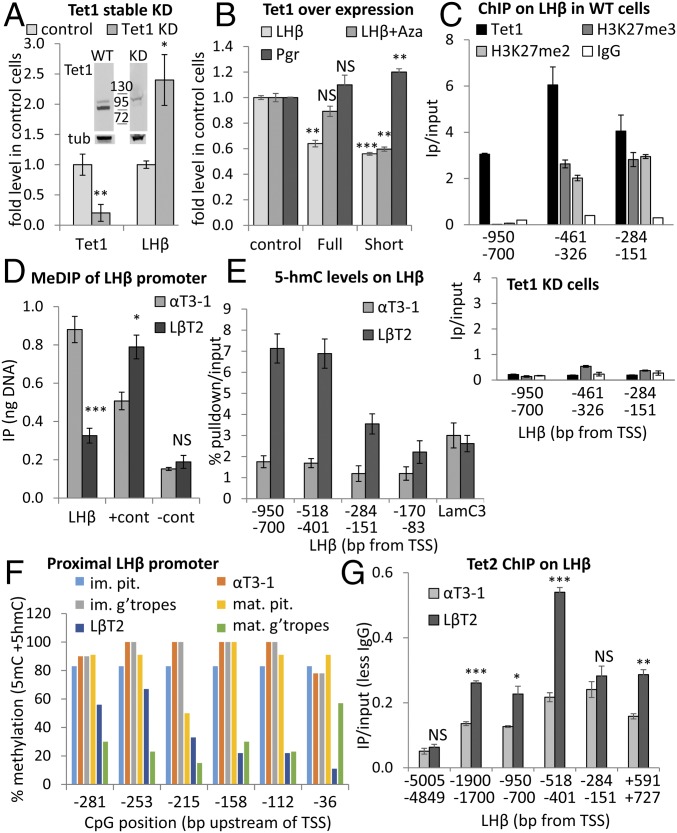
The Truncated TET1 Represses Lhb Gene Expression Regardless of DNA Methylation and Does Not Catalyze 5hmC.
To determine whether this truncated TET1 isoform regulates Lhb gene expression, we first performed stable TET1 KD, which led to an increase in Lhb mRNA levels (Fig. 5A). Conversely, over-expression of the full-length or truncated isoform repressed Lhb similarly. This contrasted with the effect on the Pgr gene, whose expression was significantly, albeit marginally, increased by the truncated but not the full-length isoform (Fig. 5B), in accordance with reports that the catalytic domain (in both isoforms) but not the full-length TET1 decreases Pgr methylation (24). Binding of the TET1 CXXC domain was reported to be affected by DNA methylation (24), so we also treated the cells with 5-Aza-dC before over-expressing each isoform. The effect of the full-length TET1 on Lhb was lost after Aza treatment, whereas the truncated isoform was still inhibitory (Fig. 5B), indicting distinct mechanisms of recruitment.
Fig. 5.
The truncated TET1 represses Lhb gene expression, regardless of DNA methylation, and does not catalyze 5hmC. (A) Tet1 and Lhb mRNA in αT3-1 cells after TET1 KD; data were analyzed and presented as before (n = 3–4). Western blot shows TET1 KD (lanes are from one blot, localized exactly as shown). (B) The canonical (full) or the truncated Tet1 isoform (short) were over-expressed in αT3-1 cells, some of which were treated with Aza for 48 h, and levels of Lhb or Pgr mRNA were measured, and are shown relative to control cells and analyzed as before (n = 2–3). (C) ChIP for TET1, H3K27me2, and me3 at the Lhb promoter in WT αT3-1 cells (Top) or for TET1 and H3K27me3 in TET1 KD cells (Bottom) as in Fig. 2. (D) MeDIP analysis at the Lhb gene promoter, calculated and presented as in Fig. 3B (n = 6). (E) 5hmC DNA at the Lhb promoter, with Lamc3 as positive control, shown relative to input (n = 4–6). (F) BS analysis of the Lhb promoter in gonadotropes from immature and mature mice, nongonadotrope pituitary cells (pit), and the gonadotrope cell lines. Data presented as in Fig. 3F; n = 6–20 clones from individual or pooled mice pituitaries and n = 9–10 clones for each cell line. (G) ChIP for TET2 upstream of Lhb, presented as in Fig. 3C (n = 3). Statistical analysis compared levels at each region between the cell lines.
We went on to evaluate by ChIP whether this repressive effect on Lhb is direct, and saw that the endogenous TET1 is associated with the Lhb promoter (Fig. 5C). Moreover, ChIP for H3K27 di- and trimethylation revealed that both modifications are enriched in the proximal region of the promoter, but not in the upstream region; this was absent in the TET1 KD cells (we tested only K27me3; Fig. 5C). Thus, the endogenous truncated TET1 appears to repress expression of the Lhb gene directly, possibly involving recruitment of histone H3K27 methyltransferases.
We next determined the methylation status of the Lhb promoter and saw a much higher level of 5mC in αT3-1 cells than in the LβT2 cells (Fig. 5D). We also analyzed levels of 5hmC, which, conversely, were higher in LβT2 cells than in αT3-1 cells, especially at the more distal region (Fig. 5E). Thus, the binding of TET1 in immature gonadotrope precursor cells concurs with a region that is 5mC- but not 5hmC-methylated, whereas the 5hmC in mature cells is clearly not the result of TET1. The methylation status of the proximal region was further confirmed in primary cells from immature and mature mice by BS analysis. Despite the inherently heterogeneous gonadotrope populations, the Lhb proximal promoter was clearly the least methylated in the differentiated gonadotrope cells of the mature mice, as in the LβT2 cells, whereas the nongonadotrope pituitary cells, immature primary gonadotropes, and cell line were all methylated to a similarly high level (Fig. 5F).
Given that the truncated TET1 was clearly not responsible for 5hmC upstream of the Lhb gene promoter in the LβT2 cells, we next examined whether TET2 might be responsible for this modification. ChIP revealed that TET2 is indeed bound at significantly higher levels in the LβT2 cells than the αT3-1 cells, over the entire upstream region and particularly upstream of −401 bp from the TSS (Fig. 5G), correlating with the most enriched region of the 5hmC. Thus, in cells in which the Lhb gene is expressed, its promoter is 5hmC-modified, apparently catalyzed by TET2, whereas, in partially differentiated cells, TET1 has a predominant and repressive role on this gene to which it recruits other repressors, and does not catalyze 5hmC (Fig. S5). Expression of Lhb is thus dependent on down-regulation of Tet1 in these cells.
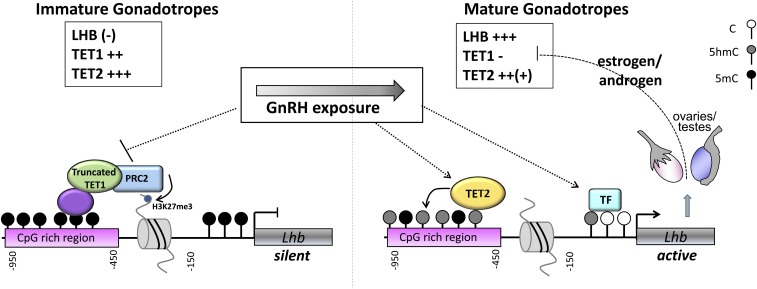
Fig. S5.
Working model for the regulation of the Lhb gene by TET1 during gonadotrope differentiation (Discussion). (Left) In immature, poorly differentiated gonadotropes (as in αT3-1 gonadotrope-precursor cells and proliferating gonadotropes in immature or gonadectomized mice), Tet1 and Tet2 are readily expressed, but Lhb expression is at low or very low levels (minor differences between the cell models noted in brackets). In these cells, the upstream CpG-rich region of Lhb is methylated (5mC) and associated with the truncated TET1 isoform, which represses Lhb expression, possibly involving recruitment of the methyl transferase-containing PRC2 complex, which catalyzes the repressive H3K27me3. With differentiation (Right), the levels of TET1 decrease, facilitated by exposure to GnRH, which inhibits Tet1 expression, leading to a decrease in the ratio of TET1:TET2, which allows TET2 to bind the Lhb promoter. TET2 catalyzes the hydroxymethylation (5hmC) of the CpG-rich region, whereas the region closest to the TSS is hydroxymethylated and/or demethylated to allow transcription, further facilitated by independent effects of GnRH via the activation of various gene-specific transcription factors (TFs). This state is further strengthened by the repressive effect of the steroids produced in the active gonads, which maintain repression on Tet1 in the sexually mature animal.
Discussion
We have shown here that a truncated TET 1 isoform directly represses expression of the Lhb gene, necessitating its down-regulation for final gonadotrope differentiation, and have also revealed the means of its regulation. Inhibition of this TET1 isoform by GnRH during the prepubertal period facilitates Lhb gene expression, but might also comprise an additional mechanism to curb proliferation of the gonadotrope precursor cell population (25). Certainly, the elevated Tet1 expression in proliferating immature gonadotropes suggests that it plays a role in establishing this population of cells during development, which would at least partially explain the effect of its KO on fertility (11, 12). Subsequent to its repression by increasing GnRH levels, there is further transcriptional inhibition of this Tet1 isoform by the gonadal steroids, in keeping with previous reports on the repressive effects of a synthetic estrogen on Tet1 in the uterus (26); both would affect Tet1 in gonadotropes but not thyrotropes. Notably, the Lhb gene appeared more sensitive to the increase in TET1 in females than in males, possibly relating to the differing phenotypes in the Tet1 KO mice, although this is likely also the result of the complex hormonal interplay in the gonadotrope during the estrous cycle. Our findings thus not only extend a recent report that this isoform is expressed in various adult somatic tissues (27), but also place it in a physiological context, describing its role and regulation in the gonadotrope, through an alternative promoter as well as a distal enhancer.
The TET1-mediated repression of Lhb is direct, with TET1 binding the CpG-rich region on the more distal part of the promoter, to which it appears to recruit histone-modifying enzymes responsible for repressive modifications. Such effects have been reported in ESCs, and TET1-bound promoters were seen to be occupied by the polycomb repression complex 2 (PRC2), leading to the suggestion that TET1 facilitates PRC2 binding, associates with the Sin3A complex, and/or may help recruit the MBD3/NuRD complex (21, 28, 29). Although this seems a likely mechanism of TET1-mediated repression of the Lhb gene, the lack of CXXC domain in this isoform suggests some differences in binding of the TET1 to the DNA, and possibly also its function. Indeed, the region of the Lhb gene promoter bound by the truncated TET1 is clearly 5mC- but not 5hmC-methylated, and hypomethylation by Aza had no effect on its repressive activity. This contrasted with the full-length TET1, whose repressive activity on Lhb was lost in the hypomethylated cells, presumably because of competition from the increased number of unmethylated regions (24).
Apart from this difference in sensitivity to the methylation status of the DNA, the truncated TET1 appears to be catalytically inactive, certainly in the current context of the Lhb gene promoter, but also apparently in other differentiated cells in which this truncated isoform is the dominant form, according to our PCR analysis and also ENCODE and CAGE data for histone modifications and TSSs in a large number of tissues. Unlike in ESCs, in which TET1, PRC2, and H3K27me3 colocalize with 5hmC, in these differentiated tissues, colocalization with 5hmC is not apparent (30). This was clarified in a recent study (27) reporting that, aside from the CXXC domain, the N terminus of the full-length TET1 also contains a domain that promotes the demethylation activity, which is thus reduced in the short isoform. Therefore, the truncated isoform clearly harbors characteristics distinct from those of the canonical full-length isoform, not only in the way it is recruited, but also in its function. In fact, the Lhb gene is 5hmC-methylated only when Tet1 expression is reduced, and TET2 then binds the same region to catalyze the modification, facilitating Lhb expression.
Although we have shown that this truncated Tet1 isoform is regulated through an alternative proximal promoter regulated by gonadal steroids and PKA, we also describe an upstream enhancer. Methylation of this enhancer plays a crucial role in determining Tet1 expression, but it can be protected by TET2. This distal region is enriched at its 5′ end with a high ratio of HK4me1 to HK4me3, whereas the CpGI core is enriched with HK4me3, characteristic of unmethylated CpGIs (31). Moreover, the region adjacent to that enriched with H3K4me1 is transcribed, and the transcript levels were found to vary in accordance with basal, but not GnRH-regulated, expression of Tet1 mRNA. The 3C assay showed that this region interacts physically with the functional Tet1 TSS, all of which suggest that this distal region acts as a transcriptional enhancer for the truncated Tet1 isoform.
Another group recently reported that, in stem cells, this distal CpGI comprises one of two alternative TSSs for a longer Tet1 transcript that includes exon 1, and these are used differently at distinct stages of development (32). However, we were unable to amplify any fragment by using various forward primers targeting the CpGI 1 and reverse primers targeting exon 1 [termed exon 2 in their paper (32)], confirming that this gene is indeed regulated very differently through cell differentiation and development. In support of these distinct regulatory mechanisms, the aforementioned study (32) showed enrichment of H3K27 acetylation at this upstream region (i.e., 1a and vicinity), which was completely lacking in the gonadotropes, and does not appear in other differentiated tissues, in accordance with the lack of exon 1 expression. This distinct regulation of Tet1 in stem cells is perhaps not surprising given that its expression is activated by pluripotent factors in these cells (9, 10), and the absence of such factors in differentiated cells would necessitate alternative regulatory mechanisms and the use of additional cis-elements.
The present study supports the idea that distinct genomic regions can be used differently at various stages of development or in different tissues (23) and extends the possibilities of a particular sequence being transcribed as part of the primary transcript in some tissues, or as an eRNA from a functional enhancer in others. We have shown that this truncated Tet1 isoform, which is more common in differentiated tissues than the canonical ESC form, is regulated by methylation at two genomic regions, as well as a common kinase signaling pathway and gonadal steroids, opening the way for studies of its regulation in diverse contexts. These findings therefore have broad implications in understanding the regulation of Tet1 expression in other tissues, especially hormonally regulated cancers. Regardless of its precise role in proliferating gonadotrope precursor cells, we have shown that down-regulation of this Tet1 isoform is essential for Lhb expression and therefore also in gonadotrope cells completing differentiation and subsequently acquiring reproductive competence.
Materials and Methods
Mice, Cells, and Culture.
αT3-1 and LβT2 murine gonadotrope-derived cells were cultured and treated (17, 18) as detailed in SI Materials and Methods. Pituitaries were extracted from GRIC-GFP or GRIC-tdTomato mice; gonadotropes were enriched by FACS (as in refs. 33,34), and some were cultured for 24 h before treatment (as in ref. 25 and SI Materials and Methods). Ovariectomy and orchiectomy (www.iacuc.ucsf.edu/Policies/Gonadectomy.doc) and other animal experiments were performed after protocol approval by the Institutional Animal Care and Use Committee of Technion – Israel Institute of Technology and the State of Saarland review board, and according to institutional animal care and use committee guidelines.
RNA Extraction, Real-Time PCR, and 5′RACE.
RNA was isolated with TRIzol, DNase I digested, and cDNA synthesized (High Capacity cDNA reverse transcription kit; Applied Biosystems). qPCR was performed with PerfeCTa SYBR Green FastMix (Quanta) with primers listed in Table S1. Calculation of amplicon levels and their analysis are detailed in SI Materials and Methods. RACE reactions were as previously described (18).
Table S1.
Primers
| Primer no. | Gene | Sequence |
| RT-PCR | ||
| 76 | GAPDH +51F | ATGGTGAAGGTCGGTGTGAA |
| 77 | GAPDH +280R | TCCTGGAAGATGGTGATGGG |
| 350 | Lhb +179F | CTAGCATGGTCCGAGTACTG |
| 351 | Lhb +301R | CTGAGGGCTACAGGAAAGGA |
| 184 | RPLP0 +196F | GCGACCTGGAAGTCCAACTA |
| 185 | RPLP0 +296R | ATCTGCTTGGAGCCCACAT |
| 326 | Tet1 exon 11F | GAGCCTGTTCCTCGATGTGG |
| 327 | Tet1 exon 11R | CAAACCCACCTGAGGCTGTT |
| 790 | Tet1 exon 1F | TCCTAGGACAGGCCTTTAGT |
| 791 | Tet1 exon 1R | CTATCCATATCCATAGCCAC |
| 792 | Tet1 exon 3F | CTGGAACAAGTGGTAGCCAT |
| 793 | Tet1(5′RACE) exon 3R | GGTCTTTTCTCACCGAATGA |
| 798 | Tet1 exon 2F | GTTACAAAAGAAAACAAGAGGC |
| 800 | Tet1 exon 1.5F | GGAGGCTTCCTCTGCAGAA |
| 801 | Tet1 exon 1.5R | TGCATAAATAAAGATGGAGGG |
| 804 | Tet1 exon 2R | CTTTAAAACTTTGGGCTTCTTT |
| 799 | Tet1(5′RACE nested) exon 3R | TTCAAGGGGATGTGTGATCAA |
| 292 | Tet2 +4248F | ACTTCTCTGCTCATTCCCACAGA |
| 293 | Tet2 +4347R | TTAGCTCCGACTTCTCGATTGTC |
| 294 | Tet3 +3393F | GAGCACGCCAGAGAAGATCAA |
| 295 | Tet3 +3492R | CAGGCTTTGCTGGGACAATC |
| 219 | Fshb +637F | GCTGGAGAGCAATCTGCTGCCA |
| 220 | Fshb +795R | TATTGGGCCGAGCTGGGTCCTTA |
| 35 | Cga +97F | ATGGATTACTACAGAAAATATGCAG |
| 36 | Cga +196R | CCTGAATAATAAAGTCTCCATCAGG |
| 1194 | PGR F | CAGAAAGGGGTTGTCCCCAG |
| 1195 | PGR R | TTCCGGAAATTCCACAGCCA |
| ChIP/MeDIP/5hmC real-time PCR | ||
| 21 | Lhb +591F | GTCCGAGTACTGCCGGCT |
| 351 | Lhb +727R | CTGAGGGCTACAGGAAAGGA |
| 661 | Lhb −107F | CCCAAAGAGATTAGTGTGTCTAG |
| 662 | Lhb −33R | TGCGGGTTGTGGGGGTGGCA |
| 363 | Lhb −83R | GCTTGGGTAACCTAGACACT |
| 260 | Lhb −284F | GACGTTTACTCCAGCAATCTGG |
| 181 | Lhb −151R | GTGTCCCAGTGAATTGGCCTCA |
| 370 | Lhb −170F | CACTGGGACACTGGAGCTA |
| 366 | Lhb −518F | CTTACTTCCAGAGTTCCTCC |
| 367 | Lhb −401R | GCACCACCGTCTCCCAGCCC |
| 55 | Lhb −950F | ATCTGAACTCTGCTGCACTT |
| 56 | Lhb −700R | TGTACTGGGCAAATTCGGTC |
| 59 | Lhb −1900F | GAATCTTTGGGCTCCCTTC |
| 60 | Lhb −1700R | CAGGCTCCTTCCTGCAGTGC |
| 727 | Lhb −4849R | GGCAGTTCCCAAGCCTGGA |
| 596 | Lhb −5005F | GCTCTCAGAATGCAAGGCTA |
| 396 | Lamc3 +89F | ACATGGGCTCTTGCTACGAC |
| 397 | Lamc3 +316R | GGCTGTGGAAGTCTGTGAGG |
| 415 | GAPDH −226F | GGAAGCAGCATTCAGGTCTC |
| 416 | GAPDH −148R | CAGGATAGGACTCAGGGAATACAG |
| 808 | Tet1 −24,439F | GTAAGTACACCATAGCTGTCCT |
| 809 | Tet1 −24,314R | AGTGCCCACTGCTCTTCCTAA |
| 831 | Tet1 −22,274F | GTCACCTGGATGACCCGACT |
| 1154 | Tet1 −22,098F | AGGAAGGAGTGGACAGGAGAA |
| 1155 | Tet1 −22,037F | TCGCATTTTGCCTCTCGCTCA |
| 626 | Tet1 −21,911F | GTACCCGCGCACTCCGCTC |
| 1168 | Tet1 −21,800F | CTGCCTCTTCTACGGGAACATTCG |
| 627 | Tet1 −21,782R | GTTCCCGTAGAAGAGGCAGG |
| 628 | Tet1 −21,491F | GGATCTCGATCGGGCTAGG |
| 830 | Tet1 −21,420R | GCGCGGAGTTGACTCGCAC |
| 629 | Tet1 −21,361R | CGCGAGCATTGTTAGTCACC |
| 785 | Tet1 −21,187F | GGCGATAGCTTTTGCCAGCGA |
| 783 | Tet1 −20,943F | GACTGGTTCTGGGATCCTGGA |
| 784 | Tet1 −20,840R | GCTCCCGGAGCTCACGTGCA |
| 787 | Tet1 −19,349F | GAGTTAACTATTGCTCTCAGATT |
| 788 | Tet1 −19,216R | CAGGAAGGATGTGTCCAGAAG |
| 643 | Tet1 −14,665F | GGCAACACCTCCAGATTCTC |
| 770 | Tet1 −14,572R | CTCGAGGGCTGCAGAGTAAGTAAAGA |
| 843 | Tet1 −191F | GCTAAGGGAGACTCCTCCAT |
| 853 | Tet1 −2R | AAGCTTTCCCAGACAAAGGATCTCCT |
| 839 | Tet1 −53F | GCCTGTGTCCTGTGGTGAGA |
| 838 | Tet1 +68R | CCACAAAGAGTGCAGCCAAT |
| 1178 | Tet1 (−1900) F | TTGCTCTATACAGCTATGGCT |
| 1179 | Tet1 (−1600) R | CTGCCCTGCCCTGTACAAA |
| 1180 | Tet1 (−1580) F | TGGAGGATGGTGTAAAGAGAA |
| 1181 | Tet1 (−1300) R | TTGGCAGGATTGCCTGGCTGC |
| 1182 | Tet1 (−1280) F | ATCCTGCAAATGTACTGCAG |
| 1183 | Tet1 (−1000) R | ACTGCACCCATACTCAGTGA |
| 1184 | Tet1 (−950) F | TCAGGTGTGTGTGTGTGTGT |
| 1185 | Tet1 (−700) R | GATGACCTTCAGTGCCCAG |
| 1249 | Tet1 (−720) F | GTTTCTCTGGGCACTGAAGG |
| 1250 | Tet1 (−400) R | ATTGGCGATATTGAAGGCAG |
| 1189 | Tet1 (−330) F | GCACAATACCTTCCAGGGTT |
| 1190 | Tet1 (−1) R | TCCCAGACAAAGGATCTCCT |
| 173 | H19F (positive MeDIP control) | GCATGGTCCTCAAATTCTGCA |
| 174 | H19R (positive MeDIP control) | GCATCTGAACGCCCCAATTA |
| 624 | Tet1 -22,569F (negative MeDIP control) | GTGTAAAGCCACAAGCCAGT |
| 625 | Tet1 -22,464R (negative MeDIP control) | GCATATCCCAAGGTAAGATAG |
| 790 | Tet1 exon 1F (AR ChIP negative control) | TCCTAGGACAGGCCTTTAGT |
| 791 | Tet1 exon 1R(AR ChIP negative control) | CTATCCATATCCATAGCCAC |
| 1366 | Tubb3 ARE1 forward (AR ChIP positive control) | TGGCCCCCAGAACAGAAG |
| 1367 | Tubb3 ARE1 reverse (AR ChIP positive control) | TGGTGTTCCCACTCTGTACAATG |
| BS conversion PCR | ||
| 1158 | Tet1 −296F | GTATAATATTTTTTAGGGTTGTTGTG |
| 1159 | Tet1-42R | CAAAACACAAACAACCACAAACTAC |
| 691 | Tet1 −21,929F | GGGTTGATTTGGTTGGGAGTATT |
| 692 | Tet1 −21,617R | CCTACACCAATCCTTAAACAAACT |
| 693 | Tet1 −21,639F | GTTTGTTTAAGGATTGGTGTAGGTTT |
| 694 | Tet1 −21,312R | ATCCTAACCAAAACTAACTAAACTT |
| Chromatin conformation assay | ||
| 843 | Tet1 −191 | GCTAAGGGAGACTCCTCCAT |
| 842 | Tet1 −221 | GACCTGCCCTGGTGCCACC |
| 878 | Tet1 −3416 | GCATGTGCAGGACTGGAGAGA |
| 845 | Tet1 −14991 | GATTCACTATGTAGCTATGGATG |
| 844 | Tet1 −15011 | TGTAGCCCAGACTGGCCTCA |
| 873 | Tet1 −15122 | GAATCGCCATGTACATGCTGG |
| 874 | Tet1 −18282 | GGAACTTGCTCTGTAGACCAGG |
| 875 | Tet1 −19592 | GATATGCACCCTCCACTGCTG |
| 851 | Tet1 −21581 | CAGAGGCAGTTTAGGGTGAGC |
| 850 | Tet1 −21680 | GCCTCAGGCTCCAAAGTTGC |
| 852 | Tet1 −22485 | GCTATCTTACCTTGGGATATGC |
| 876 | Tet1 −24521 | GAGCCATTTCGCCTGCCCAA |
Methylation Analysis.
Sonicated denatured genomic DNA was precipitated with mouse anti-5mC (Diagenode) and sheep anti-mouse IgG Dynabeads (Invitrogen). After washing and proteinase K digestion, the DNA was extracted and precipitated before resuspension in water for qPCR. BS analysis used the EZ-DNA Methylation-Direct Kit (Zymo) and Red Load Taq Master (Larova). After purification (PCR purification kit; Qiagen) and cloning into pGEM-T-Easy (Promega), inserts from at least six random clones were sequenced and analyzed by Quantification Tool for Methylation Analysis software (quma.cdb.riken.jp/). Hydroxymethyl Collector Kit (Active Motif) was used to label the 5hmC, and the eluted DNA was analyzed by qPCR as detailed earlier (primers in Table S1).
ChIP.
ChIP was carried out after cross-linking (as in ref. 18) with the following antibodies: TET1 and ESR1 (ab-191698 and ab-32063; Abcam); TET2 and AR (sc-136926 and sc-816 X; Santa Cruz Biotechnology); histones and RNAPII as in ref. 18; or IgG as control. The DNA was purified and regions amplified by qPCR from input and immunoprecipitated (IP) samples as detailed earlier (primers in Table S1).
Protein Extraction.
Western analysis was carried out as described previously (35) with TET1 (09–872; Millipore), TET2 (ab94580; Abcam), H3 (ab1791; Abcam), and secondary (sc-2004; Santa Cruz Biotechnology) antibodies.
Chromatin Conformation Capture Assay.
Using a Dpn2-digested library of ligated fragments from αT3-1 cells (as in ref. 18), PCR used nested primers at the exon 1.5 or exon 1 TSS and various upstream primers, as in Fig. S3 and Table S1. Amplicons were resolved by electrophoresis and sequenced or measured by qPCR relative to standards comprising the same cloned chimeric fragment.
Microscopy.
Formaldehyde-fixed frozen pituitaries were sectioned at 14 µm and processed (as in ref. 32) before incubation with primary antibody [TET1 (ab191698; Abcam) at 1:500 or CGA (NIDDKD) at 1:1,500] overnight, and then secondary antibody (1:500; 706-166-148; Jackson Labs) for 2 h before application of Hoechst (1:10,000) for 7 min, rinsing, and covering.
Statistical Analysis.
Data are from at least three independent experiments, combined or shown as a representative, as mean ± SEM. The n values represent biological repeats transfected or treated and measured independently. Statistical analysis was performed by Student’s t test (two-tailed), with significant differences at P ≤ 0.05, or Fisher’s exact test for the BS analysis.
SI Materials and Methods
Cell Culture and Transfections.
The αT3-1 murine gonadotrope-derived cells represent immature, partially differentiated cells and express the gonadotropin α-subunit (Cga), as well as the GnRH receptor, but barely express the Lhb and Fshb genes, whereas the LβT2 cells represent more fully differentiated gonadotropes that express also these genes, albeit Lhb at notably higher levels than Fshb (15, 17). Both were gifts from Pamela Mellon, University of California, San Diego. The cell lines were cultured and treated with GnRH (100 nM, 8 h), H89 (13.5 μM 1 h before GnRH), PMA (10 nM, 6 h), or forskolin (10 μM, 4 h), or E2 or DHT (both 10 nM, 48 h in charcoal-stripped FCS). Treatment with 5-aza-deoxycitydine was for 48 h, and renewed every 24 h (all hormones and chemicals from Sigma-Aldrich). Transient transfections were carried out at 60% confluence using GenePORTER (Genelantis) or PolyJet (SignaGen Laboratories) reagent according to the manufacturers' instructions. Following transfection, cells were incubated for 24–48 h, and fresh serum medium was replaced after 24 h. Stable transfections were carried out as reported previously (18), with linearized pSUPERGFP/neo plasmid (Oligoengine) containing the shRNA sequence (9), and selection with G418 (Sigma Aldrich) was maintained throughout.
For TET1 over-expression, the truncated isoform was amplified from the full-length cDNA plasmid (in pCMV-entry; primers: Tet1 F, GATGGATCCATGGCCCCAAATCAGAGTCCATG; R, GATGTTTAAACCTTATCGTCGTCATCC) and inserted into the EGFP-C2 (Clontech), such that both isoforms are expressed from the same promoter and are of similar sizes.
The dominant PKA construct was a gift from Stan McKnight, University of Washington, Seattle.
RNA Extraction, Reverse Transcription and Real-Time PCR, and 5′RACE.
RNA was isolated using TRIzol and up to 2 µg RNA subjected to 10 µL DNase I (Invitrogen) before cDNA synthesis using the High Capacity cDNA reverse-transcription kit (Applied Biosystems). Mouse ESC RNA was extracted from CGR8 cells, a gift from Daniel Aberdam, Technion, Haifa, Israel, and mammary tissues, brain, placenta, liver, and heart from the GRIC-tdTomato mice. Real-time PCR was carried out using the PerfeCTa SYBR Green FastMix (Quanta) with primers listed in Table S1. Amplicon levels were analyzed in triplicate and quantified relative to a standard curve comprising cDNA or genomic DNA, and values were normalized to levels of the housekeeping gene (Rplp0 or Gapdh). Reactions were 3 min at 95 °C, then 40 cycles of 10 s at 95 °C and 30 s at 60 °C with addition of a melt curve step of 10 s at 95 °C, and increments of 0.5 °C every 5 s between 65 °C and 95 °C. 5′RACE was carried out on αT3-1 cell cDNA as previously described (18) using nested primers shown in Table S1, and the start site was identified by sequencing.
Isolation of Genomic DNA for Methylation Analysis.
Cells (106–107) were harvested in PBS solution, centrifuged (1,700 × g, 5 min), and resuspended in 300 µL TE and 300 µL MeDIP lysis buffer (20 mM Tris⋅HCl, pH 8, 4 mM EDTA, 20 mM NaCl, 1% SDS) containing 10 µL proteinase K (20 mg/mL stock). The samples were sonicated (2 × 15 s, amplitude 33%) and incubated at 55 °C for at least 5 h. Samples were extracted with 1 volume phenol and centrifuged (21,000 × g, 10 min at 4 °C) before collection of the aqueous phase, which was mixed with 1 volume chloroform and centrifuged as before. The aqueous phase was collected, mixed with two volumes EtOH containing 75 mM Na acetate, pH 5.2 (1.2 mL), and incubated at −20 °C for 1 h. Samples were then centrifuged (21,000 × g, 15 min at 4 °C), and the pellet was washed with 70% EtOH. The DNA was resuspended in TE with 20 µg/mL RNase A and incubated for 1 h at 37 °C. For MeDIP or hydroxymethyl collector experiments, the DNA was diluted with 300 µL TE and again sonicated (10 × 15 s, amplitude 33%) in ice water. Gel electrophoresis confirmed the fragment size (100–500 bp). The sonicated DNA was precipitated with 400 mM NaCl, two volumes EtOH, and 1 µL of glycogen blue at −20 °C for 1 h. The DNA was then centrifuged (21,000 × g, 15 min at 4 °C), and the pellet was washed with 70% EtOH before drying and resuspension in 20 µL TE. For BS conversion, after the RNase A treatment, the DNA was precipitated with EtOH without sonication.
Acknowledgments
We thank Pamela Mellon for the gonadotrope cell lines and Stan McKnight for the dominant-negative PKA. This research was supported by German Research Foundation (Deutsche Forschungsgemeinschaft) Grants BO 1743/7-1 (to U.B. and P.M.) and SFB894 (to U.B.), and Israel Science Foundation Grant 840/12 (to P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.F.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704393114/-/DCSupplemental.
References
- 1.Zhang H, et al. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto H, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y-F, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song C-X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neri F, et al. TET1 is controlled by pluripotency-associated factors in ESCs and downmodulated by PRC2 in differentiated cells and tissues. Nucleic Acids Res. 2015;43:6814–6826. doi: 10.1093/nar/gkv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melamed P, Kadir MN, Wijeweera A, Seah S. Transcription of gonadotropin beta subunit genes involves cross-talk between the transcription factors and co-regulators that mediate actions of the regulatory hormones. Mol Cell Endocrinol. 2006;252:167–183. doi: 10.1016/j.mce.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Lim S, et al. Distinct mechanisms involving diverse histone deacetylases repress expression of the two gonadotropin beta-subunit genes in immature gonadotropes, and their actions are overcome by gonadotropin-releasing hormone. Mol Cell Biol. 2007;27:4105–4120. doi: 10.1128/MCB.00248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melamed P. Histone deacetylases and repression of the gonadotropin genes. Trends Endocrinol Metab. 2008;19:25–31. doi: 10.1016/j.tem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Wijeweera A, et al. Gonadotropin gene transcription is activated by menin-mediated effects on the chromatin. Biochim Biophys Acta. 2015;1849:328–341. doi: 10.1016/j.bbagrm.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Pnueli L, Rudnizky S, Yosefzon Y, Melamed P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin α-subunit gene. Proc Natl Acad Sci USA. 2015;112:4369–4374. doi: 10.1073/pnas.1414841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudnizky S, et al. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat Commun. 2016;7:12958. doi: 10.1038/ncomms12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith PF, Keefer DA. Immunocytochemical and ultrastructural identification of mitotic cells in the pituitary gland of ovariectomized rats. J Reprod Fertil. 1982;66:383–388. doi: 10.1530/jrf.0.0660383. [DOI] [PubMed] [Google Scholar]
- 21.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 23.Melamed P, Yosefzon Y, Rudnizky S, Pnueli L. Transcriptional enhancers: Transcription, function and flexibility. Transcription. 2016;7:26–31. doi: 10.1080/21541264.2015.1128517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin C, et al. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014;42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savulescu D, et al. Gonadotropin-releasing hormone-regulated prohibitin mediates apoptosis of the gonadotrope cells. Mol Endocrinol. 2013;27:1856–1870. doi: 10.1210/me.2013-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jefferson WN, et al. Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Mol Endocrinol. 2013;27:1666–1677. doi: 10.1210/me.2013-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, et al. Isoform switch of TET1 regulates DNA demethylation and mouse development. Mol Cell. 2016;64:1062–1073. doi: 10.1016/j.molcel.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 28.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neri F, et al. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013;14:R91. doi: 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohni A, et al. Dynamic switching of active promoter and enhancer domains regulates Tet1 and Tet2 expression during cell state transitions between pluripotency and differentiation. Mol Cell Biol. 2015;35:1026–1042. doi: 10.1128/MCB.01172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen S, et al. Functional characterization of genetically labeled gonadotropes. Endocrinology. 2008;149:2701–2711. doi: 10.1210/en.2007-1502. [DOI] [PubMed] [Google Scholar]
- 34.Hoivik EA, et al. DNA methylation of intronic enhancers directs tissue-specific expression of steroidogenic factor 1/adrenal 4 binding protein (SF-1/Ad4BP) Endocrinology. 2011;152:2100–2112. doi: 10.1210/en.2010-1305. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, Lawson MA, Melamed P. A proteomic comparison of immature and mature mouse gonadotrophs reveals novel differentially expressed nuclear proteins that regulate gonadotropin gene transcription and RNA splicing. Biol Reprod. 2008;79:546–561. doi: 10.1095/biolreprod.108.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]