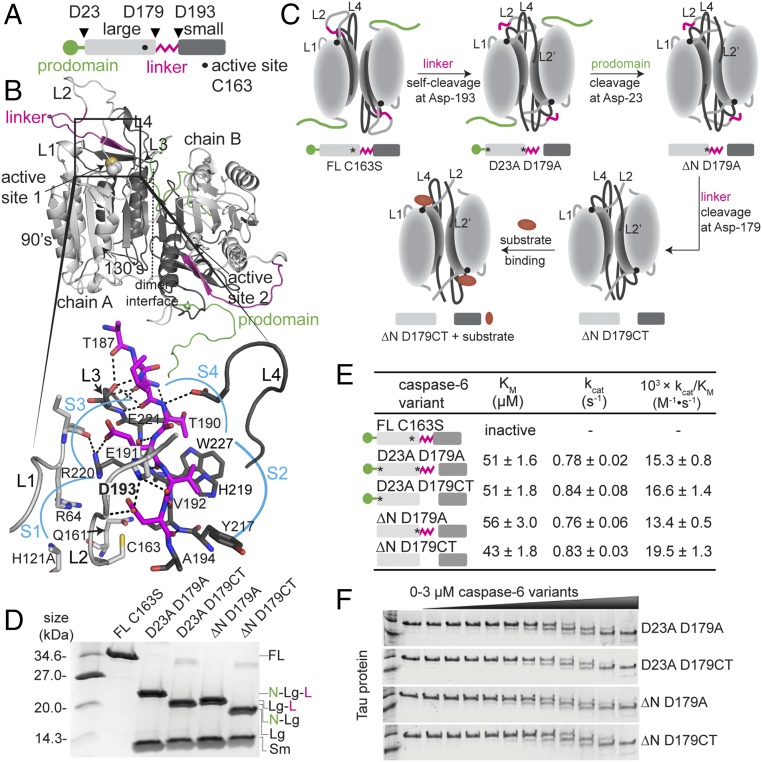
Fig. 1.
Caspase-6 maturation-state variants. (A) Linear cartoon of procaspase-6 illustrating the prodomain (green), large subunit (light gray), intersubunit linker (magenta), and small subunit (dark gray). The active site C163 is denoted by a dot. Triangles indicate proteolytic cleavage sites. (B) Model of full-length (FL) procaspase-6 zymogen colored as in A was generated based on zymogen structures 4IYR and 3NR2 with missing regions modeled de novo. Inset shows details of binding interactions of the intersubunit linker residues (magenta) binding into the S1–S4 subsites. (C) Cartoon of expected conformational changes in caspase-6 constructs representing various points along the zymogen maturation (cleavage) pathway colored as in A, with substrate as an ellipsoid (red). FL indicates FL uncleaved procaspase-6 zymogen. The C163S catalytic site substitution renders caspase-6 inactive and incapable of self-activation. ΔN indicates removal of the N-terminal prodomain (1–23). D23A substitution renders caspase-6 uncleavable after the prodomain. D179A prevents cleavage of the intersubunit linker. D179CT constructs have a stop codon inserted after residue 179 to allow expression of a constitutively two-chain caspase-6, mimicking native cleavage at D179. The asterisk designates D to A mutation in the proteolytic cleavage site in caspase-6. (D) Maturation-state caspase-6 variants purified from overexpression in E. coli. (E) Catalytic properties of caspase-6 maturation-state variants. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. (F) Cleavage of Tau-383 protein (3 μM) is not impacted by the maturation state of caspase-6, tested at increasing concentrations (0–3 μM).