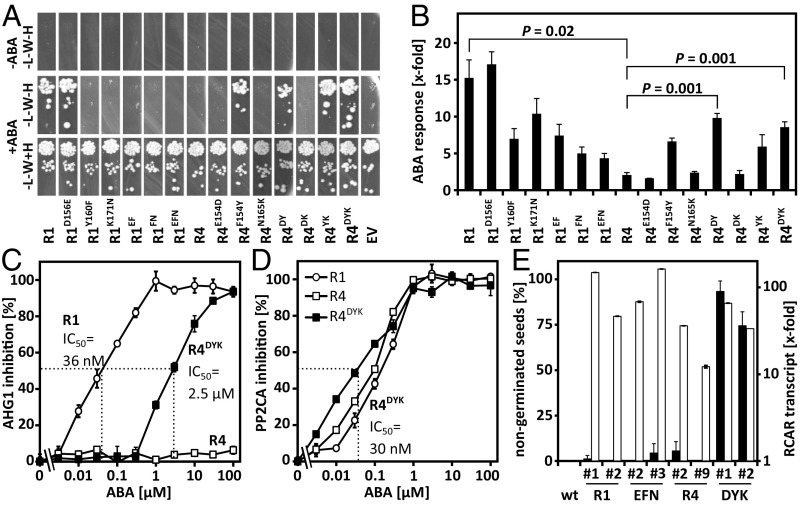
Fig. 5.
Specificity determinants of RCAR1 and RCAR4 for AHG1 regulation and interaction. (A) Protein interaction analysis of AHG1 and variants of RCAR1 and RCAR4 in yeast shows the critical role of a Tyr residue in RCAR1 and ABA for AHG1 binding. Histidine autotrophic growth of yeast (−H) spotted as a 10-fold dilution series on media deficient in Leu (−L) and Trp (−W) indicates RCAR–AHG1 interaction. The RCAR variants were modified at the amino acid residues highlighted in yellow in Fig. 4B. (B) Rescue of AHG1-imposed inhibition of ABA signaling by different RCAR variants in the presence of 10 µM exogenous ABA. The analysis was performed in protoplasts with the RCAR variants as mentioned in Fig. 4A (triplicates, mean ± SD). (C and D) Biochemical analysis of PP2C regulation by ABA in the presence of RCAR1 (open circle), RCAR4 (open squares), and the modified RCAR4DYK (black squares). (C) The phosphatase activity of AHG1 (2 µM) and (D) PP2CA (0.4 µM) was assayed in the presence of twofold molar excess of RCAR protein and different ABA concentrations as indicated. The IC50 values for ABA of the RCAR-AHG1 regulation are depicted. AHG1 is not regulated by RCAR4 (open squares) even at 0.1 mM ABA (mean ± SD). (E) Analysis of RCAR-overexpressing Arabidopsis lines for seed germination and RCAR overexpression. The percentage of nongerminated seeds was assessed in independently generated lines overexpressing RCAR1/RCAR1-EFN variant and RCAR4/RCAR4-DYK variant in comparison with the parental line Col-0 after 2 d of stratification at 4 °C and 3 d of incubation at room temperature (filled bars, n > 90 per data point, mean ± SD). The RCAR1 and RCAR4 transcript levels were determined in seedlings and expressed relative to WT (open bars).