Significance
We describe an approach to blocking protein–protein interactions in living cells and use it to probe the earliest stages of epigenetic regulation in stem cell differentiation. We describe a computationally designed protein that tightly binds EED and disrupts PRC2 function in both cancer and stem cells. Expression of the binder at different stem cell stages identifies the first critical repressive H3K27me3 mark in embryonic development.
Keywords: polycomb repressive complex, Rosetta protein design, human embryonic stem cell, human early development, epigenetics
Abstract
The polycomb repressive complex 2 (PRC2) histone methyltransferase plays a central role in epigenetic regulation in development and in cancer, and hence to interrogate its role in a specific developmental transition, methods are needed for disrupting function of the complex with high temporal and spatial precision. The catalytic and substrate recognition functions of PRC2 are coupled by binding of the N-terminal helix of the Ezh2 methylase to an extended groove on the EED trimethyl lysine binding subunit. Disrupting PRC2 function can in principle be achieved by blocking this single interaction, but there are few approaches for blocking specific protein–protein interactions in living cells and organisms. Here, we describe the computational design of proteins that bind to the EZH2 interaction site on EED with subnanomolar affinity in vitro and form tight and specific complexes with EED in living cells. Induction of the EED binding proteins abolishes H3K27 methylation in human embryonic stem cells (hESCs) and at all but the earliest stage blocks self-renewal, pinpointing the first critical repressive H3K27me3 marks in development.
Preimplantation (ground and naïve) embryonic stem cells (ESCs) with high developmental potential and postimplantation (primed) ESCs with more restricted potential have been stabilized in mice and in humans and have distinctive metabolic and epigenetic landscapes (1). The functional relevance of H3K27me3 marks in human ESCs (hESCs) is unclear. In mouse during blastocyst formation, PRC2 complex-dependent repression of CDX2 and GATA3 is essential for the ICM lineage (2), and reprogramming assays have revealed an essential function for PRC2 in acquisition of pluripotency (3). Furthermore, pluripotency depends on the chromatin-based silencing of developmental gene expression (4). However, mouse ground state ESCs are able to self-renew and express stem cell markers without PRC2 (5, 6). The contradiction between these studies raises the possibility that different stages of pluripotency have different requirements for PRC2-dependent repressive histone marks as observed for DNA methylation (7); consistent with this, H3K27me3 levels are quite dynamic in early preimplantation embryos (8). To investigate the function of PRC2 in hESCs, we used small molecules that inhibit EZH2 action and CRISPR methods (SI Appendix, Fig. S1A). Five-day treatments with Astemizole (0.5 µM) and EPZ-6438 (2.5–5 µM) (9, 10) resulted in dramatic morphological changes in Elf1 hESCs (SI Appendix, Fig. S1A). Recovered CRISPR EED null cells (SI Appendix, Fig. S1G) lacked the Oct4 stem cell marker, and clonal EED null hESC lines did not self-renew: All of the single clonal lines generated from the CRISPR pools expressed EED (SI Appendix, Fig. S1H). While the small molecule and CRISPR data are suggestive of a role for PRC2 in maintaining hESC pluripotency, the long time required for a phenotype in the former case and the incomplete penetrance in a pool and difficulty in maintaining clonal homozygous null lines in the latter case complicate mechanistic investigation.
To enable conditional disruption of PRC2 function, we set out to design EED-binding proteins that disrupt the EED–EZH2 interaction. We aimed to stabilize the EED-binding segment of EZH2 as with “stapled peptide” approaches but using a rigid protein scaffold instead of a chemical cross-link (11). To incorporate the EED interacting domain of Ezh2 (12, 13) into a larger host protein (Fig. 1 A and B and SI Appendix, Fig. S2), we used Rosetta to identify backbone segments in the protein structure databank (Protein Data Bank, PDB) closely matching the EED-binding segment of EZH2 (residues 40–68) and superimposed these segments onto the EZH2 peptide in the EZH2–EED structure, carrying along the remainder of the protein scaffold containing the matching segment (14). The scaffold was then redesigned to maximize binding affinity with EED. This approach has the advantages over stapled peptide approaches that (i) the functional conformation can be more precisely stabilized by extensive interactions with the rest of the scaffold, (ii) the protein can make additional interactions across the interface outside of the peptide segment to further increase binding affinity, and (iii) the inhibitor can be genetically encoded and expressed in an inducible manner.
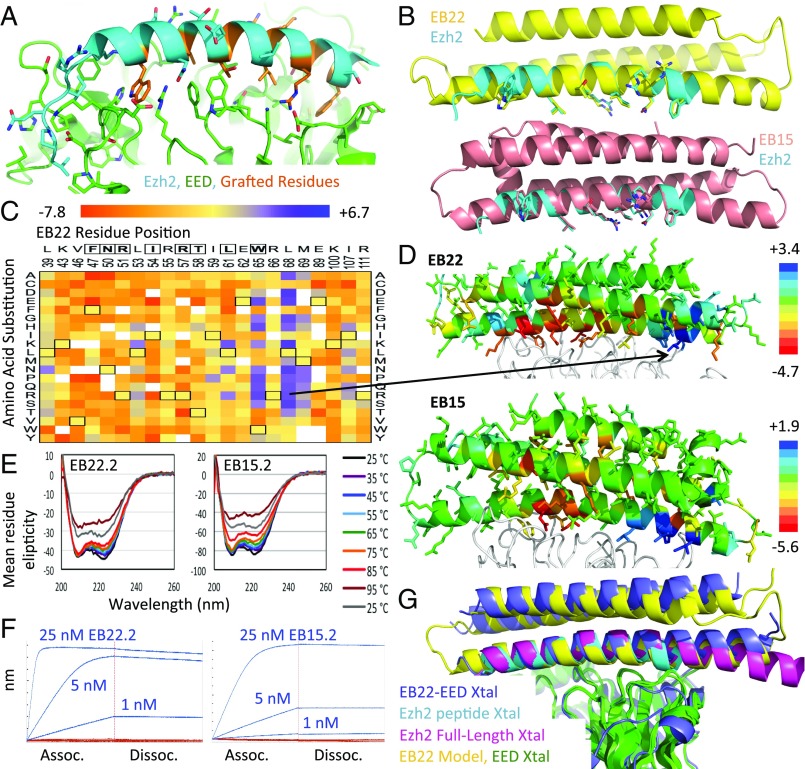
Fig. 1.
Design of EED binding proteins. (A) Crystal structure of Ezh2 N terminus (cyan) bound to EED (green) (PDB ID code 2qxv) with grafted residues highlighted in orange. (B) Models of EB22 (yellow, Top) and EB15 (salmon, Bottom) superimposed onto the Ezh2 peptide (cyan) with grafted residues shown as sticks. (C) Heat map of Log2 enrichments of EB22 interface position substitutions after one round of sorting. Grafted residues are boxed at the top of the heat map, while wild-type residues are boxed within the heat map. (D) Models of EED (white ribbon) bound to EB22 (Top) and EB15 (Bottom), each colored according to the median Log2 enrichment at each position after one round of sorting. The most enriched EB22 point mutation is indicated in the model by a black arrow. (E) CD thermal melts of EB22.2 (Left) or EB15.2 (Right). (F, Left) BLI traces of 1, 5, or 25 nM EB22.2 (blue) or EB22.2NC (red) bound to EED. (F, Right) As in Left, but for EB15.2 and EB15.2NC. (G) EB22–EED crystal structure (blue) or design model (yellow) superimposed (by aligning the EED molecules) onto crystal structures of Ezh2 peptide-EED (cyan) and full-length Ezh2–EED (magenta).
Genes encoding the designs with the highest predicted EED-binding affinity were synthesized, and EED binding was assessed using yeast surface display (15) and FACS. Deep mutational scanning (16) of the two highest affinity designs, EB15 and EB22, revealed that the interacting residues in the N-terminal half of the EED binding surfaces were strictly conserved, suggesting that EB15 and EB22 bind EED as in the design model (Fig. 1 C and D and SI Appendix, Fig. S4). New polar interactions are introduced in the C-terminal half, complementary to charges on EED; the most enriched point variants of each protein are hereafter referred to as EB15.2 and EB22.2. EB15 and EB22 core residues were conserved even at less stringent selection conditions, consistent with the design models (Fig. 1 C and D and SI Appendix, Figs. S4–S6).
Circular dichroism of EB22, EB22.2, and EB15.2 expressed and purified from Escherichia coli showed they are well folded and have melting temperatures of >95 °C (Fig. 1E and SI Appendix, Fig. S7). EED-binding affinities of EB15.2 and EB22.2 were subnanomolar by both biolayer interferometry (BLI) and on-yeast titrations (15) (Fig. 1F and SI Appendix, Tables S1–S3)—much stronger than those of EZH2 and previous EZH2 analog stapled peptides (11, 12). On-rates for EB15.2 (1.60E05 ± 3.82E03 M–1·s–1) and EB22.2 (7.79E05 ± 1.87E04 M–1·s–1) were improved by two orders of magnitude relative to EZH2 (2.17E03 ± 7.31E01 M–1·s–1) as measured by BLI (Fig. 1F and SI Appendix, Table S1). Stabilization of the helical EZH2 fragment in the active binding conformation via our scaffolding approach likely preorders the binding epitope, leading to a substantial increase in observed on-rates. Negative control variants of EB15.2 and EB22.2 each lacking two key interface residues (EB15.2NC and EB22.2NC) do not bind to soluble EED (Fig. 1F and SI Appendix, Tables S2 and S3).
The crystal structure of EB22 bound to EED shows the binding mode is very similar to that of the design model except that the N terminus of the binding helix is displaced 2 Å away from the surface of EED and translated 1 Å along its axis toward its C-terminal end; similar shifts are observed in recent crystal structures of EED bound to Ezh2 (Fig. 1G and SI Appendix, Fig. S8). Rosetta modeling suggests that EB22 Leu-68 may desolvate the phenolic hydroxyl group of Tyr-154, while the EB22.2 Arg-68 substitution may make near-ideal hydrogen bonds to this hydroxyl (SI Appendix, Fig. S9). Ezh1 likely binds to EED at the same site as Ezh2 (SI Appendix, Fig. S10), and hence our designs likely compete with both Ezh1 and Ezh2 binding to EED.
We expressed EB22.2 and the negative control EB22.2NC as GFP fusion proteins under the control of a tetracycline-inducible expression promoter in cancer cells (Fig. 2A). Western blot detection and mass spectrometry analysis after immunoprecipitation with GFP antibody revealed specific binding of EB22.2 to EED (Fig. 2 A and B and SI Appendix, Table S6). Disruption of the EED–EZH2 interaction by EB22.2 reduces EZH2 levels (Fig. 2C and SI Appendix, Fig. S11); qRT-PCR analysis of EZH2 mRNA level and Western blot detection of EZH2 after cycloheximide chase suggests that EZH2 loss is due to protein turnover (Fig. 2C and SI Appendix, Fig. S12), recapitulating previous observation (17). Expression of EB22.2 reduced interactions of EED with SUZ12, RBBP4, and AEBP2 (Fig. 2D, Right), which appears to increase the accessibility of EED antibody to EED and increases the amount of immunoprecipitated EED. EB22.2 decreased the coelution of EED with EZH2, SUZ12, RBBP4, and AEBP2 in sucrose gradient centrifugation (Fig. 2E, fractions 7–13), suggesting dissociation of EED from the other PRC2 components. Unlike EZH2, SUZ12, and RBBP4, a portion of AEBP2 elutes in fractions 3–6, where most of EED elutes, suggesting residual interactions between EED and AEBP2, independent of the EZH2–EED interaction (18) (Fig. 2D).
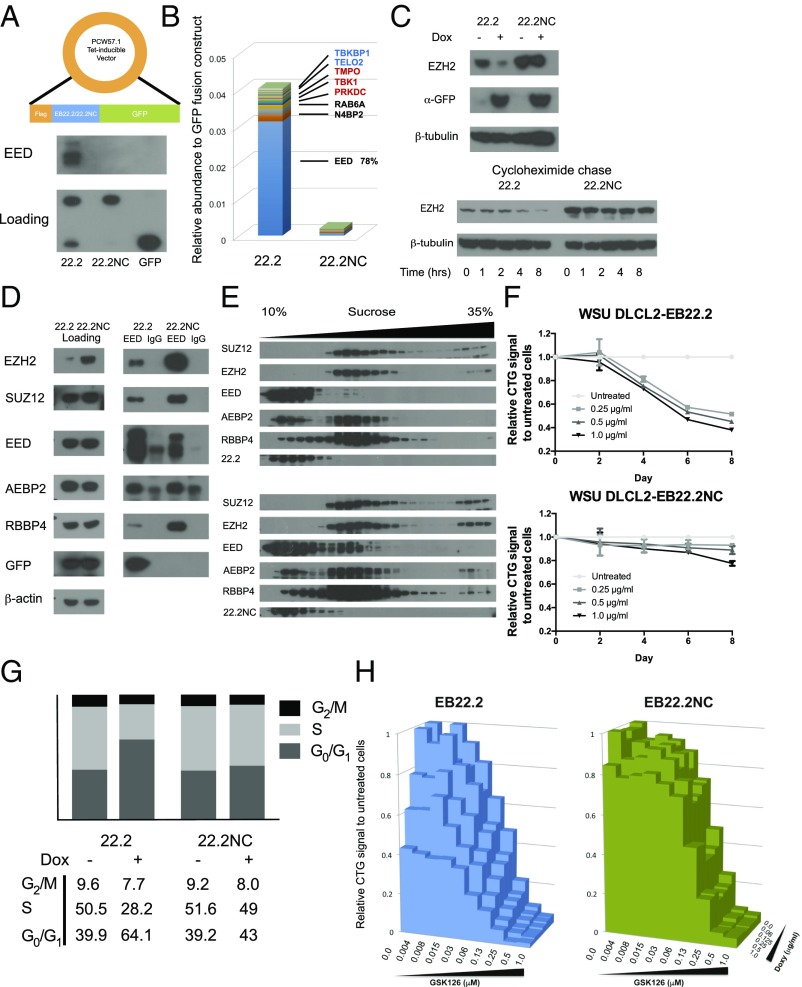
Fig. 2.
Functional validation of the designed protein. (A–H) EB22.2/EB22.2NC were expressed using tet-inducible vectors at 0.5 µg/mL dox induction for 48 h unless otherwise indicated. (A, Top) EB22.2/22.2NC expression construct. (A, Bottom) Co-IP and detection of EED by EB22.2 from K562 cell lysates. (B) Co-IP by GFP antibody and mass spectrometry analysis from K562 cells. Red, confirmed in BioGrid database; blue, unconfirmed in BioGrid database. (C, Top) Detection of EZH2 in K562 cells expressing or not expressing EB22.2/EB22.NC. (C, Bottom) Detection of EZH2 in EB22.2/EB22.2NC expressing K562 cells after exposure to cycloheximide. (D) Detection of each PRC2 component in EB22.2/EB22.2NC expressing K562 cells before (Left) or after (Right) co-IP by EED antibody. EED, co-IP by EED antibody; IgG, co-IP by rabbit IgG control to assess nonspecific binding. (E) Western blot of PRC2 components fractionated by sucrose gradient centrifugation from K562 cell lysate expressing EB22.2 (Top) or EB22.2NC (Bottom). (F) Proliferation of WSU-DLCL2 cells expressing EB22.2 (Top) and EB22.2NC (Bottom). CTG, cell titer glo. Data represent mean data ± SEM for independent experiments performed in triplicate. (G) Cell cycle analysis of WSU-DLCL2 cells expressing EB22.2/EB22.2NC. BrdU-7AAD staining and flow cytometry analysis data are in SI Appendix, Fig. S17. Expression of EB22.2/EB22.2NC was induced for 4 d. (H) Proliferation of WSU-DLCL2 simultaneously treated with various concentrations of GSK126 and doxycycline for EB22.2 expression. CTG signal of each data point is normalized by the CTG value at Dox 0.0 µg/mL and GSK126 0.0 µM. Each data point represents the mean for independent experiments performed in duplicate. Expression of EB22.2/ EB22.2NC was induced for 8 d.
We investigated the effects of EB22.2 on the proliferation of PRC2-dependent diffuse large B-cell lymphoma (DLBCL) cell lines (19, 20). EB22.2 dissociated the PRC2 complex and decreased the proliferation of both WSU-DLCL2 and Pfeiffer cells (Fig. 2F and SI Appendix, Figs. S13 and S14) not by inducing apoptosis (SI Appendix, Figs. S15 and S16) but by perturbing the cell cycle, as observed with the hydrocarbon stapled peptide of Ezh2 (11) (Fig. 2G and SI Appendix, Figs. S17 and S18). EB22.2 is collaborative with the small-molecule inhibitors: Exposure of WSU-DLCL2 cells to both EB22.2 and GSK126 reduced proliferation compared with exposure to either EB22.2 or GSK126 (Fig. 2H and SI Appendix, Figs. S19 and S20).
To investigate the role of EZH2–EED in shaping the hESC epigenetic landscape, we introduced doxycycline-inducible EED binder, EB22.2 (EB22.2-NLS-GFP; EB22.2-Flag), and the negative control EB22.2NC (EB22.2NC-NLS-GFP; EB22.2NC-Flag) constructs in the AAVS1 site in pluripotent cells [WTC human induced pluripotent stem cells (21) and hESCs (WIBR3, Elf1); Fig. 3 A and K and SI Appendix, Figs. S21–S23]. We grew these cells in different growth conditions to stabilize primed and naïve states (primed, TeSR; naïve, 2iL-I-F, 4/5iLA). Endogenous EED was the most abundant protein identified by mass spectrometry in EB22.2 pulldowns (Fig. 3 B and C and SI Appendix, Fig. S21 B–J), while the controls (EB22.2NC or GFP) did not coimmunoprecipitate EED (SI Appendix, Fig. S21 E–G and Table S7).
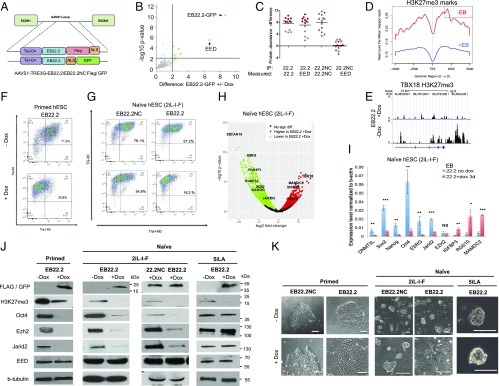
Fig. 3.
PRC2 is required for primed and intermediate but not naïve 5iLA hESC pluripotency. (A) EED-binder-AAVS1-TRE3G Tet-inducible construct models; Flag or GFP: EED-binder (EB22.2) = EB22.2-Flag-NLS or EB22.2-NLS-GFP, negative control (EB22.2NC) = EB22.2NC-Flag-NLS or EB22.2NC-NLS-GFP. (B) EB22.2-GFP binds to EED; co-IP from Elf1 cells was quantified by mass spectrometry. Proteins associated with black lines represent a cutoff of 0.01 (P value) and a difference (log2 transformed) greater than two ±Dox. (C) Protein co-IP from plus or minus dox-treated cells measured by mass spectrometry. n = 4–6 technical measurements from three independent cell lines. Gray, 0.5 µg/mL Dox; red, 2 µg/mL Dox. EB22.2, filled circle; EB22.2NC-FLAG-GFP, filled inverted triangle; EB22.2NC, filled square; GFP, filled triangle. (D) Three days of treatment with Dox in Elf1 EB22.2-FLAG cells decreases H3K27me3 marks. H3K27me3 reads mapped 5 kb around transcription start sites (TSSs) of 374 genes that are expressed twofold higher in +Dox and have H3K27me3 peaks in −Dox were plotted for ChIP-seq data. (E) Refseq gene position of TBX18 (embryonic development transcription factor) and H3K27me3 profiles detected by ChIP in Elf1-EB22.2-FLAG treated or not with Dox for 3 d. (F) Primed Elf1 EB22.2-FLAG lose stem cell markers Tra-1–60 and TG-30 using flow cytometry. Cells were grown for 5 d in TeSR followed by 48 h of Dox induction (2 µg/mL). (G) Naïve Elf1 EB22.2-FLAG but not EB22.2NC-FLAG shows important reduction of stem cell markers Tra-1–60 and TG-30 using flow cytometry. Cells were grown for 3 d in condition media (CM) + 2IL-I-F under Dox induction (2 µg/mL). (H) Volcano plot representation of mRNA expression in naïve hESCs Elf1 EB22.2-FLAG treated or not for 3 d with Dox (2 µg/mL). Genes with greater than twofold change and expressed above 10 reads per million are colored. Genes with known roles in regulating development and pluripotency as well as bivalent genes in Elf1 are labeled. (I) qRT-PCR analysis of most differentiated expressed genes between naive Elf1 EB22.2-FLAG 2iL-I-F cells treated or not with Dox for 3 d (n = 3, SEM; *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t test). (J) Primed and naïve 2iL-I-F EB22.2 leads to PRC2 disruption and Oct4 down-regulation, but not in EB22.2-naïve 5iLA or NC22.2 using immunoblot analysis. Primed EB22.2s were grown in TeSR media for 30 d and were either induced for 5 d (+Dox) or not induced at all (−Dox). Naïve 2iL-I-F EB22.2s or NC22.2s were grown in CM+ 2iL-I-F for 3 d in the presence of Dox (+Dox) or not (−Dox). Naïve 5iLA EB22.2s were grown in CM+ 5iLA for 3 d in the presence of Dox (+Dox) or not (−Dox). (K) Morphology of primed WTC, naïve Elf1 2iL-I-F, and naïve Elf1 5iLA EB22.2 and NC22.2 after 5 d of Dox treatment (primed) or 3 d of Dox treatment (naïve). (Scale bars, 100 µm and 50 µm, respectively.)
Cells expressing EED binder showed a significant genome-wide reduction in H3K27me3 repressive epigenetic marks in the promoter regions of gene loci (Fig. 3 D and E and SI Appendix, Fig. S21 K–Q and Tables S13 and S14). In the primed hESC conditions, induction of EB22.2, but not EB22.2NC, changed colony morphology and sharply reduced H3K27me3 and EZH2 levels and pluripotency markers Tra1-60 and Oct4 (Fig. 3 F, J, and K and SI Appendix, Fig. S22 A and B). Primed human PSCs evidently require the EED/EZH2 interaction to maintain stemness; a PRC2 requirement at this stage in mouse ESC has also been suggested (22). Consistent with this, EZH2 chemical inhibitors reduced H3K27me3 marks, stem cell colony morphology, and Oct4 expression in both human and mouse primed ESCs (SI Appendix, Fig. S23 D–G).
To determine if PRC2 is required earlier in hESCs at the naive and naïve-to-primed transition, we induced EB22.2 and EB22.2NC expression in naïve hESC(2iL-I-F). After EB22.2 but not EB22.2NC expression, hESC(2iL-I-F) colonies cultured on Matrigel flattened out and lost stem cell morphology (Fig. 3K and SI Appendix, Figs. S21D and S23A). Flow cytometry analysis revealed a significant reduction in surface expression of the stem cell marker Tra-1–60 in EB22.2 but not in control EB22.2NC samples (Fig. 3G). RNA-seq analysis and qPCR validation revealed a dramatic down-regulation of stem cell markers and up-regulation of differentiation genes (Fig. 3 H and I). The levels of PRC2 proteins EZH2 and Jarid2 were also reduced (Fig. 3J and SI Appendix, Fig. S23C), but EZH2 RNA levels were unchanged, suggesting instability of the EZH2 protein (Fig. 3I). The major pluripotency marker Oct4 was dramatically down-regulated both on the RNA and protein levels with EB22.2 but not EB22.2NC expression (Fig. 3 I and J and SI Appendix, Fig. S23B), further indicating that the 2iL-I-F naïve cells expressing EED binder protein failed to maintain self-renewing pluripotency.
To analyze the PRC2 requirement in the naïve-to-primed transition, during which H3K27me3 and IDO1 levels increase and DNMT3L and NNMT levels decrease (SI Appendix, Fig. S22 C and D), we expressed EB22.2 in naive hESC(2iL-I-F) during the transition; this resulted in a considerable reduction of Tra1-60, suggesting loss of the stem cell fate (SI Appendix, Fig. S22 E–I). These results contrast with earlier findings of PRC2 independence of self-renewal in mouse ESCs (23). The discrepancy could reflect species differences between mouse and human or differences in stem cell developmental stage. To investigate the latter possibility, we carried out principal component analysis (PCA) of RNA-seq data. The results suggest that naïve hESC 4/5iLA or Klf2/Nanog-induced lines (24–26) are the closest to the human in vivo preimplantation blastocyst, followed by 2iL-I-F hESC (23), the 3iL (27), and the mouse ESC line (Fig. 4A).
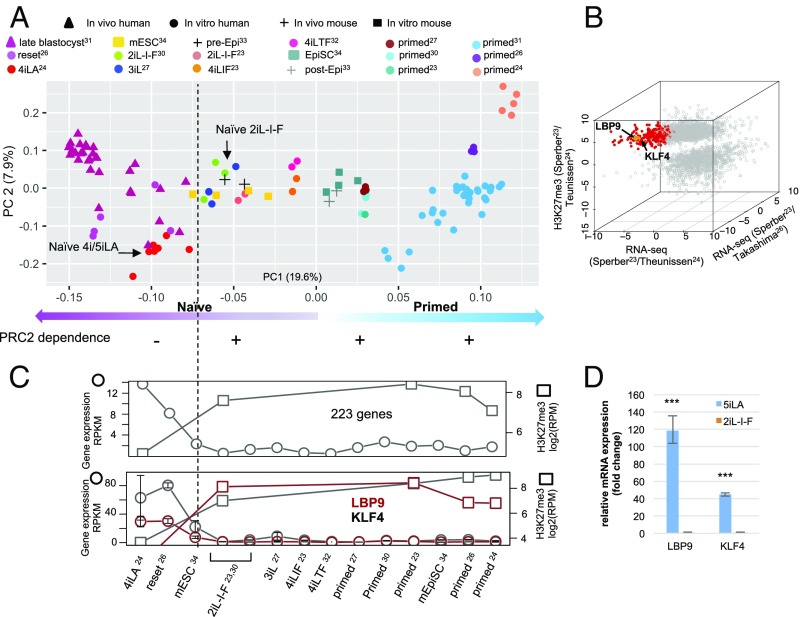
Fig. 4.
Developmental ordering of the naïve-like hESC lines based on expression signatures. (A) Naïve hESCs require PRC2 for Oct4 expression after transition from a naïve 4/5iLA to a naïve 2iL-I-F pluripotent state. PCA plot of human and mouse in vivo and in vitro pluripotent stem cell samples. RNA-seq samples from multiple studies that investigated naïve and primed pluripotency in vivo and in vitro in human (23–27, 30, 31, 32) and mouse (33, 34) are uniformly processed and aggregated in this principal component plot. PC1 is associated with naïve–primed pluripotency state. (B) 223 genes with significantly lower expression (RNAseq) in naïve 2iL-I-F compared with the naïve lines from Theunissen et al. (24) (x axis) and Takashima et al. (26) (y axis) and significantly higher H3K27me3 (z axis) in naïve 2iL-I-F compared with Theunissen et al. (24) naïve 4/5iLA are marked in red. Two genes known to regulate naïve 5iLA pluripotency, LBP9 and KLF4, are among this group of genes. (C) Gene expression and normalized H3K27me3 reads within 2 kB of TSS for 223 genes (Upper) and LBP9 and KLF4 (Lower) showed a significant increase from a 5iLA naïve (24) pluripotent state to naïve 2iL-I-F and primed hESC lines. (D) LBP9 and KLF4 are up-regulated in Elf1 5iLA compared with Elf1 2iL-I-F, as shown by qRT-PCR analysis. (n = 3, SEM; ***P < 0.001, two-tailed t test.)
To determine whether the earliest stabilized hESC stages require PRC2, we transitioned the EB22.2 hESC(2iL-I-F) line into the 5iLA naïve state. In these 5iLA naive state cells, H3K27me3 marks were down-regulated after EED binder expression, but the Oct4 expression was unaltered (Fig. 3 J and K). Similar results were obtained with the EZH2 catalytic inhibitor EPZ-6438 on naïve 4iLA hESCs with two different genetic backgrounds (WIBR3) (25): The drug reduced H3K27me3 marks, but the pluripotency marker Oct4 and the stem cell colony morphology were not affected, as seen with the EB22.2 5iLA cell line (Fig. 3 J and K and SI Appendix, Fig. S23 D–F). These data suggest that the earliest pluripotent naïve hESC state is not sensitive to PRC2 loss, narrowing the window when hESCs become PRC2 dependent to the stage between 5iLA and 2iL-I-F (Fig. 4A). We identified 223 genes with increased H3K27me3 marks and reduced expression levels in hESC 2iL-I-F versus 5iLA state (24, 26) (Fig. 4 B and C). These include the naïve state markers LBP9 and KLF4 (28, 29) (Fig. 4 B–D), which show significant down-regulation of expression and an increase of H3K27me3 marks 5 Kb around transcription start sites as pluripotent cells move from the naïve 5iLA state to the later, possibly preimplantation epiblast stage (Fig. 4). PRC2 may not be required in the naïve 5iLA state because its first targets function at this stage.
Induced expression of the designed EED inhibitor reveals distinct requirements for PRC2 in hESCs; while the naïve 5iLA state is PRC2 independent, later states require PRC2 to maintain pluripotency. Introducing biologically designed inhibitors allows the probing of cellular function with this high temporal resolution. The PRC2 dependence and RNA-seq data together suggest a consistent ordering of the mouse and hESC lines stabilized to date: 4/5iLA state (two genetic backgrounds, WIBR3 and Elf1) and mouse naïve ESC state are at an early PRC2-independent pluripotent state, and other hESCs at a slightly later, PRC2-dependent state. Our findings set the stage for deciphering the mechanistic origins of the onset of a PRC2 requirement in the exit from the early pluripotent state and the function of the newly introduced repressive H3K27me3 marks.
Supplementary Material
Acknowledgments
We thank Prof. Ware and members of the H.R.-B. laboratory for helpful discussions during this work and Jennifer Hesson for help with 5iLA hESC transition and culturing. This work is supported by NIH Grants R01GM097372, R01GM97372-03S1, and R01GM083867 and NHLBI Progenitor Cell Biology Consortium Grants U01HL099997 and U01HL099993 (to H.R.-B.); the WRF Innovation Fellowship Program (S.L. and Y.X.); NIH Grant P01GM081619 (to H.R.-B. and R.T.M.); University of Washington’s Proteomics Resource Grant UWPR95794 (NIH/NIGMS); the Defense Threat Reduction Agency (J.D.M. and D.B.); NIH Yeast Resource Center Grant GM103533 (to J.D.M., D.B., and L.T.D.); and a Claudia Adams Barr grant (to W.K.). This work is partly based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from NIH Grant P41 GM103403. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. The Structural Genomics Consortium is a registered charity (1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, the Canadian Institutes of Health Research (CIHR), Genome Canada, Ontario Genomics Institute (Grant OGI-055), Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD Grant 115766), GlaxoSmithKline, Janssen, Merck & Co., Novartis Pharma AG, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome Trust (Grant 092809/Z/10/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallographic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5WP3). The sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE102702 ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706907114/-/DCSupplemental.
References
- 1.Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 2.Saha B, et al. EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol Cell Biol. 2013;33:2691–2705. doi: 10.1128/MCB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira CF, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galonska C, Ziller MJ, Karnik R, Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao J, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 9.Kong X, et al. Astemizole arrests the proliferation of cancer cells by disrupting the EZH2-EED interaction of polycomb repressive complex 2. J Med Chem. 2014;57:9512–9521. doi: 10.1021/jm501230c. [DOI] [PubMed] [Google Scholar]
- 10.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci USA. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Z, et al. Structural basis of EZH2 recognition by EED. Structure. 2007;15:1306–1315. doi: 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Fleishman SJ, et al. RosettaScripts: A scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Correia BE, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 16.Fowler DM, Araya CL, Gerard W, Fields S. Enrich: Software for analysis of protein function by enrichment and depletion of variants. Bioinformatics. 2011;27:3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montgomery ND, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 18.Ciferri C, et al. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci USA. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandegar MA, et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 23.Sperber H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theunissen TW, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takashima Y, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan YS, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Martello G, Bertone P, Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;32:2561–2574. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naïve pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan L, et al. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 32.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 33.Boroviak T, et al. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.