Significance
We examined the effect of cell cycle arrest in the diatom Phaeodactylum tricornutum. When the cycle is disrupted in G1 phase, it leads to unbalanced growth and the accumulation of storage products, especially lipids. In contrast to nitrogen-stressed cells, however, cells arrested in G1 do not cannibalize photosynthetic proteins and show little change in photosynthetic energy conversion efficiency. This study provides insight into how intermediate metabolism is scheduled with respect to the cell cycle in a marine diatom.
Keywords: cell cycle, diatom, intermediate metabolism, unbalanced growth, flux balance analysis
Abstract
The inhibitor NU 2058 [6-(cyclohexylmethoxy)-9H-purin-2-amine] leads to G1-phase cell cycle arrest in the marine diatom, Phaeodactylum tricornutum, by binding to two cyclin-dependent kinases, CDKA1 and CDKA2. NU 2058 has no effect on photosynthetic attributes, such as Fv/Fm, chlorophyll a/cell, levels of D2 PSII subunits, or RbcL; however, cell cycle arrest leads to unbalanced growth whereby photosynthetic products that can no longer be used for cell division are redirected toward carbohydrates and triacylglycerols (TAGs). Arrested cells up-regulate most genes involved in fatty acid synthesis, including acetyl-CoA carboxylase, and three out of five putative type II diglyceride acyltransferases (DGATs), the enzymes that catalyze TAG production. Correlation of transcriptomes in arrested cells with a flux balance model for P. tricornutum predicts that reactions in the mitochondrion that supply glycerate may support TAG synthesis. Our results reveal that sources of intermediate metabolites and macromolecular sinks are tightly coupled to the cell cycle in a marine diatom, and that arresting cells in the G1 phase leads to remodeling of intermediate metabolism and unbalanced growth.
Diatoms are unicellular, eukaryotic algae that enter the fossil record approximately 150 Mya and rose to ecological prominence approximately 34 Mya in the Oligocene (1). These secondary symbionts are among the most productive and environmentally flexible algae in the contemporary oceans. They often have the capacity for extremely high carbon fixation ability and rapid growth rates compared with other planktonic microalgae, and are the dominant clade in algal blooms (2, 3).
During exponential growth in nutrient-replete conditions, ∼40% of photosynthetically fixed carbon in diatoms is incorporated into proteins (4, 5), and 15–25% is directed toward lipids, primarily in the form of algal triacylglycerols (TAGs). Absent genetic modification, TAGs increase only under certain conditions, such as nutrient starvation or stress (5–10). However, nutrient stress also interferes with fundamental functions of the cell, such as the maintenance and repair of proteins, and can lead to reduced photosynthetic rates, changes in intermediate metabolite levels, lowered growth rates, and even cessation of cell division (10–12). Thus, whereas nutrient stress leads to a remodeling of intermediate metabolism and a concomitant disruption in cell division (12), it is unclear whether the converse is true―namely, will a disruption in the cell cycle lead to remodeling of intermediate metabolism?
To understand how the cell division cycle may influence the intermediate metabolism in diatoms, we studied the effects of cell cycle arrest by inhibiting serine-threonine cyclin-dependent kinases (CDKs). CDKs interact with cyclins, a diverse family of proteins ranging in size from 35 to 90 kDa whose levels are controlled by transcription and degradation in a cell cycle-dependent manner (13). CDKs, together with cyclins, integrate metabolic cues from a network of signaling molecules to determine whether conditions are favorable for progression to subsequent phases in the cell cycle program. We hypothesize that if a cell is arrested in a specific stage in the cell cycle, then it will not be in balanced growth (by definition) and may accumulate specific end products of metabolism, such as lipids. Here, using biochemical, physiological, and systems biology approaches, we examine how a diatom remodels intermediate metabolism in response to cell cycle arrest.
Results
Selection of the CDK Inhibitor and Targets of NU 2058.
Cell cycle arrest was achieved using pharmacologic inhibitors of CDKs, which are purine analogs that target the ATP-binding pocket (i.e., the catalytic site) of these proteins. Six purine analogs were tested on the model diatom, Phaeodactylum tricornutum, to identify those that induced cell cycle arrest with minimal disruption of other basic physiological functions. The drugs tested included roscovitine, purvalanol A, purvalanol B, AT7519, Pha 79387, and NU 2058 [6-(cyclohexylmethoxy)-9H-purin-2-amine]. Of the inhibitors tested, all but Pha 79387 decreased growth rate, but only NU 2058 led to higher lipid accumulation. We eliminated the other drugs from further analysis because they either killed the cells or did not produce adequate growth arrest. NU 2058 inhibits CDK1 and CDK2 and binds to the catalytic sites in different orientations than those of roscovitine or the purvalanols (14).
We performed affinity chromatography, followed by mass spectrometry, to identify the target(s) of NU 2058 in P. tricornutum. Based on amino acid sequence similarities in the ATP-binding site in human CDK1 and CDK2, we hypothesized that NU 2058 would bind to the orthologs of these proteins in the diatom (i.e., CDKA1 and CDKA2), and indeed the NU 2058 matrix identified CDKA1 and CDKA2 as targets of NU 2058. In P. tricornutum, CDKA1 and CDKA2 have PSTAIRE and PSTALRE cyclin-binding sites, respectively (Fig. 1A). PSTAIRE is a “classic” cyclin-binding site for CDKs (15). Coimmunoprecipitation (co-IP) analysis with the PSTAIR antibody also demonstrated that PSTAIR bound to CDKA1, to CDKA2, and to a small amount of hCDK3, a hypothetical CDK.
Fig. 1.
Amino acid sequences and expression levels of P. tricornutum CDKs. (A) Partial sequences of CDKA1 and CDKA2 (P. tricornutum) and CDK1 and CDK2 (Homo sapiens). Regions highlighted in yellow are ATP-binding pockets; regions highlighted in blue are cyclin-binding regions. (B) Protein immunoblot with monoclonal anti-PSTAIR antibody. The lanes were loaded with equal amounts of protein at each time point. CDKA1 (33.11 kDa) and CDKA2 (34.12 kDa) were detected in cell lysates of dark-synchronized cultures from control cells (C) and cells with NU 2058 (N) at various time points after reillumination.
Although the precise function of CDKA1 in diatoms is unknown, transcriptomic data suggest that CDKA1 plays a key role in the G1/S transition (16). CDKA2 has been shown to play a role in the G2/M transition; it localizes to the cell division plane in precytokinetic cells, and its overexpression prolongs mitosis (17). We performed protein immunoblot analysis with the PSTAIR antibody on lysates of cells synchronized in G1 phase by prolonged darkness (24 h). On release from G1 phase by reillumination (t0), cultures were divided into controls and NU 2058 treatments. Cells increased their levels of CDKA1 and/or CDKA2 with increasing time under illumination (Fig. 1B). There were no significant differences in CDKA1 and/or CKDA2 levels between treated and untreated cells.
Growth and Physiology.
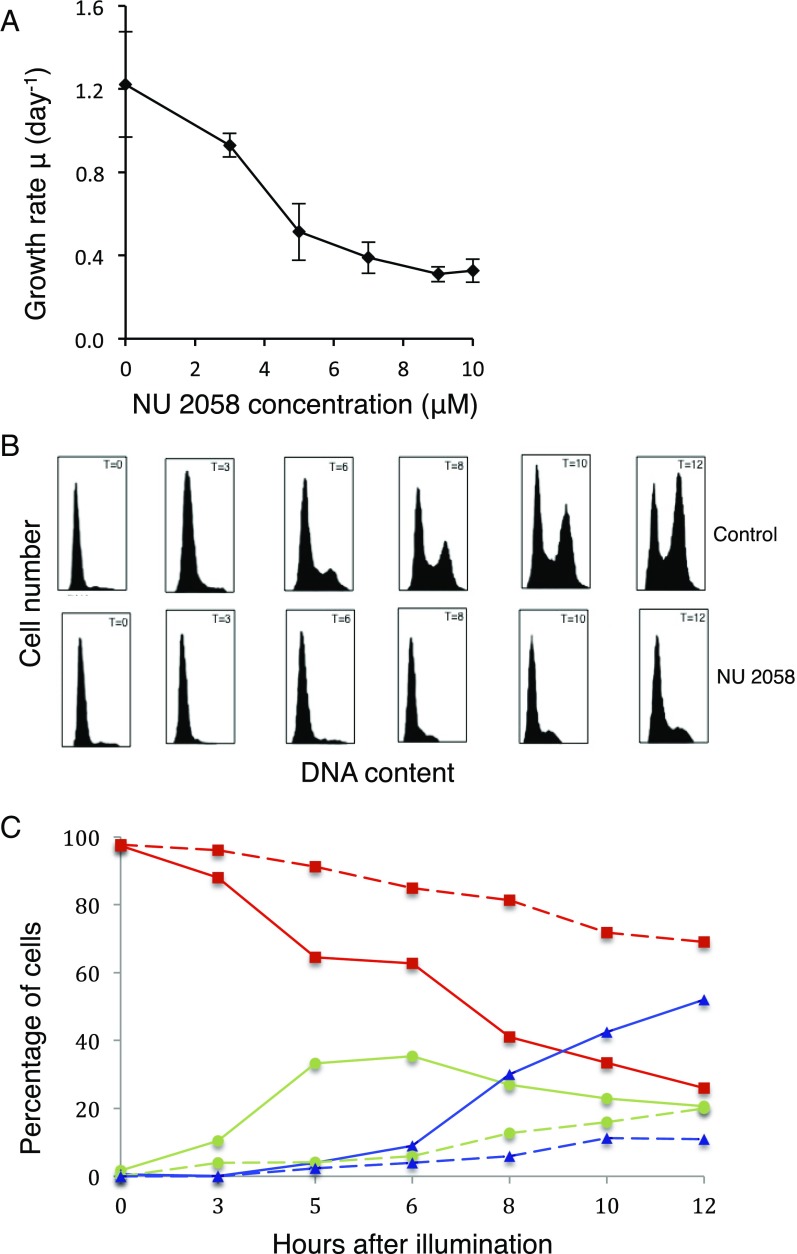
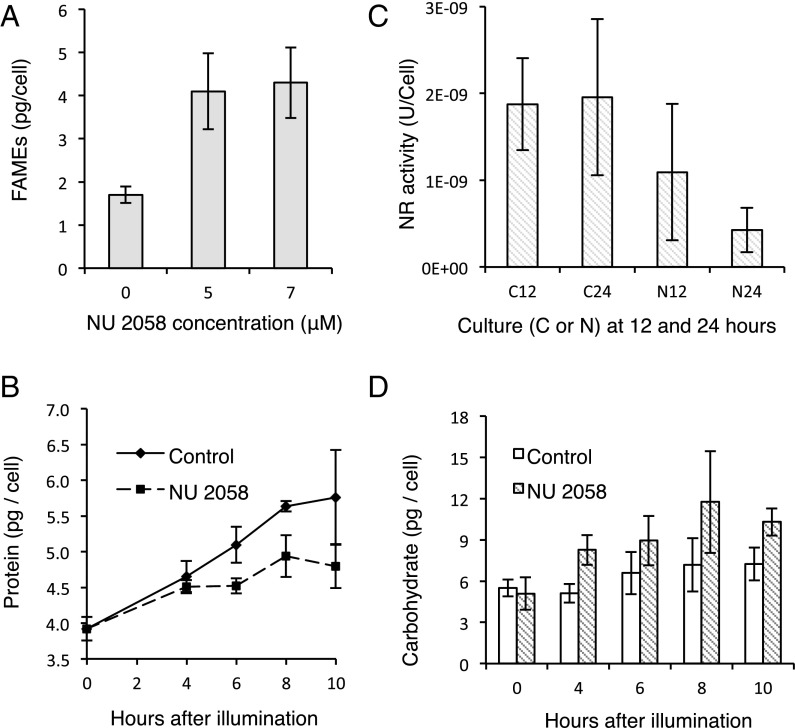
Following the addition of NU 2058 to cultures grown under continuous illumination, the growth rate of P. tricornutum decreased in a dose-dependent manner up to ∼7 μM (Fig. 2A). Meanwhile, fatty acid content [determined as fatty acid methyl ester derivatives (FAMEs)] increased with higher concentrations of NU 2058, inversely with growth rate (Fig. 3A). Cultures exposed to 7 μM NU 2058 exhibited a 2.5-fold increase in FAMEs compared with control cells. NU 2058 concentrations >7 μM did not result in significant decreases in growth rate or increases in fatty acid content; thus, we chose this concentration as the standard working dosage.
Fig. 2.
Effects of NU 2058 on growth and cell cycle in P. tricornutum. (A) Growth rate of P. tricornutum cells after 24 h of treatment with NU 2058. (B) Cell cycle analysis. Cells synchronized in G1 were placed in the light and fixed in methanol at (from left to right) 0, 3, 6, 8, 10, and 12 h after illumination. The top and bottom rows show control and treated cells, respectively. DNA was stained with propidium iodide (emission at 562–588 nm), and samples were analyzed by flow cytometry. (C) Percentages of control (solid lines) and treated (dotted lines) cells in G1 (red squares), S (green circles), and G2/M (blue triangles).
Fig. 3.
Effects of NU 2058 on physiological characteristics of P. tricornutum. (A) FAME content in cultures grown in 0, 5, or 7 μM NU 2058 for 24 h under continuous light. (B) Total protein per cell in synchronized cells at various time points after illumination. (C) Nitrate reductase (NR) activity for control (C) and treated (N) cells at 12 and 24 h after illumination. U/cell, µmol NO2− min−1 cell−1. (D) Carbohydrate per cell. Total carbohydrates were measured using glucose standards in synchronized cells at various time points after illumination. In A–D, error bars indicate SD (n = 3).
Synchronized (dark-arrested) cultures were used to better understand the effect of NU 2058 on the cell cycle. At 12 h after illumination of cultures and administration of NU 2058 (7 μM), ≈75% of cells were locked in G1 phase, compared with ≈28% of control cells (Fig. 2 B and C). At this time point, 21% of the control cells were in S phase and 52% were in G2/M phase, while approximately 14–20% of treated cells were in S phase and only 10% were in G2/M phase. Therefore, some cells with NU 2058 were able to progress into S or G2 phase, but the treated cultures did not fully recover until at least 3 d after administration of the inhibitor. Furthermore, NU 2058 administered to synchronized cells at 3, 5, or 6 h after illumination still inhibited growth, and cells were effectively locked in G1 phase of the cell cycle.
Synchronized cells were collected for macromolecules, elemental analysis, and metabolites at various time points after illumination. At t0, protein, carbohydrate, and TAGs per cell were at their lowest levels; these three macromolecular pools gradually increased under illumination. Treated cells, however, had ≈12% less (P < 0.05) protein per cell after 6 h and 8 h under illumination compared with untreated cells (Fig. 3B and Table 1). A 17% difference at 10 h was consistent with this trend, although not statistically significant (P = 0.11). The activity of the rate-limiting enzyme of nitrate assimilation, nitrate reductase (NR), was decreased by 80% in treated cells after 24 h of illumination (Fig. 3C). NU 2058-treated cells had roughly 42% higher levels of total carbohydrates by 10 h (P < 0.05) compared with untreated cells (Fig. 3D). Diatoms store polysaccharide in the form of chrysolaminarin, a linear polymer of β(1→3) and β(1→6) linked glucose units.
Table 1.
Physiological characteristics of cells in synchronized cells at 10 h after illumination
| Parameter measured | Control | NU 2058 | NU2058/control, % | NR21/WT, % |
| C, pg/cell* | 9.2 ± 0.8 | 14.01 ± 0.07 | 152 | 132 |
| N, pg/cell† | 1.68 ± 0.20 | 0.92 ± 0.21 | 55 | 105 |
| C:N, mol:mol | 6.39 | 17.76 | 278 | 128 |
| Protein, pg/cell‡ | 5.76 ± 0.66 | 4.80 ± 0.31 | 83 | 110 |
| Carbohydrate, pg/cell§ | 7.24 ± 1.19 | 10.30 ± 1.00 | 142 | 112 |
Data are mean ± 1 SD (n = 3). NR21/WT, ratio between P. tricornutum with nitrate reductase knocked down vs. WT, from Levitan et al. (12); C, carbon; N, nitrogen; C:N, carbon-to-nitrogen ratio.
P = 0.0084, Welch’s t test.
P = 0.0107.
P = 0.1105.
P = 0.0213.
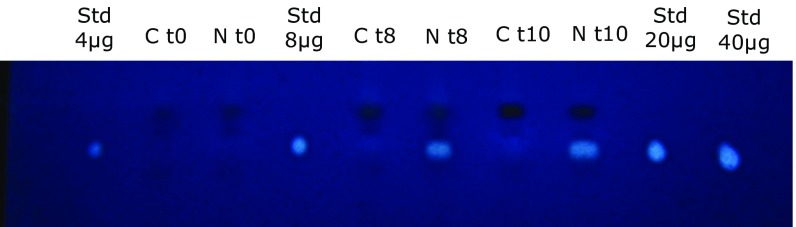
The total carbon content was approximately 52% higher, and nitrogen content was 45% lower, in treated cells compared with control cells (Table 1). Much of the increased carbon was associated with lipids, specifically TAGs. At 10 h after administration of the inhibitor, treated cells had almost twofold higher TAG levels in the light compared with untreated cells (Fig. 4). Simultaneously, however, there was a 2.8-fold decrease in intact polar lipids (IPLs), which are associated primarily with membranes (Table 2). The reduction in IPLs is similar to that observed in nitrogen-stressed cells (18).
Fig. 4.
Thin-layer chromatography showing TAG content in synchronized cells at 0, 8, and 10 h after illumination. C, control cells; N, 7 µM NU 2058-treated cells; Std, TAG standard (Sigma-Aldrich).
Table 2.
IPLs in control and NU 2058-treated cells (fmol/cell) at 10 h after illumination
| IPL | Control cells | NU 2058-treated cells |
| MGDG | 0.37 ± 0.42 | 0.05 ± 0.03 |
| DGDG | 0.55 ± 0.10 | 0.18 ± 0.03 |
| SQDG | 0.48 ± 0.15 | 0.46 ± 0.05 |
| PG | 0.37 ± 0.10 | 0.03 ± 0.02 |
| PE | 0.04 ± 0.01 | 0.00 ± 0.00 |
| PC | 0.20 ± 0.02 | 0.01 ± 0.01 |
Data are mean ± 1 SD (n = 3). MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; PG, glycerophosphoglycerols; PE, phosphatidylethanolamine; PC, phosphatidylcholine.
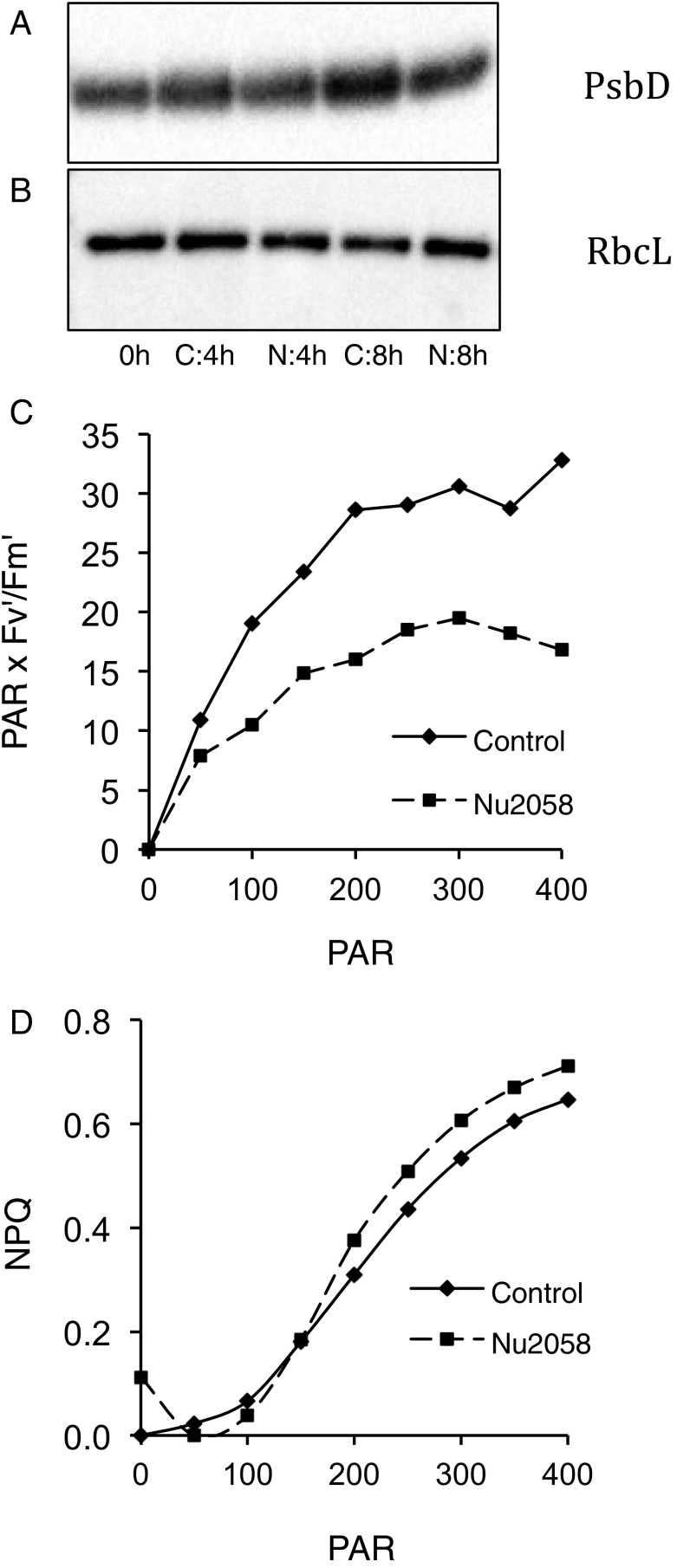
Cells exposed to NU 2058 showed no differences in maximum photochemical quantum yields of PSII (Fv/Fm), chlorophyll a (Chl a) per cell, or the in vivo optical absorption cross-section normalized to Chl a (a*) (Table 3). Moreover, there were no apparent differences in levels of PsbD (Fig. 5A) or RbcL (Fig. 5B). However, after 24 h of exposure to the inhibitor, photosynthetic electron transport rates at light saturation were ≈55% lower (Fig. 5C), and nonphotochemical quenching was 33% higher (Fig. 5D). Since a* and Chl a levels were comparable in treated and untreated cells, the quantum requirement for fatty acid biosynthesis (1/ϕFA) in treated cells was ≈60% lower than that in untreated cells (Table 3). Thus, although treated cells grew more slowly than untreated cells, they used absorbed photons more efficiently to produce lipids; that is, the flux of carbon into lipids was higher in treated cells.
Table 3.
Photosynthetic attributes of P. tricornutum at a range of NU 2058 concentrations
| NU 2058, μM | Fv/Fm | Chl a, pg/cell | a*, m2/mg Chl a | 1/ϕFA, quanta/C |
| 0 | 0.58 | 0.55 ± 0.02 | 0.006 ± 0.001 | 217 ± 13 |
| 5 | 0.60 | 0.60 ± 0.01 | 0.006 ± 0.002 | 150 ± 14 |
| 7 | 0.57 | 0.54 ± 0.03 | 0.006 ± 0.001 | 147 ± 14 |
Data are mean ± 1 SD (n = 3). Fv/Fm, maximum quantum efficiency of photochemistry; a*, optical absorption cross section of PSII normalized to Chl a; 1/ϕFA, the photon energetic requirement for incorporation of carbon into fatty acid.
Fig. 5.
Photosynthetic parameters. (A and B) Detection of PsbD (A) and RbcL (B) protein subunits by immunoblot analysis of extracts from synchronized cells at 0, 4, and 8 h after illumination. C, control cells; N, NU 2058-treated cells. Lanes were loaded on an equal protein basis (2 µg). (C and D) Electron transport rates (PAR × Fv′/Fm′) (C) and nonphotochemical quenching (NPQ) (D) in cells treated and untreated with NU 2058 after 24 h under continuous illumination. PAR, photosynthetically active radiation. Error bars indicate SD (n = 3).
Metabolite Profiles.
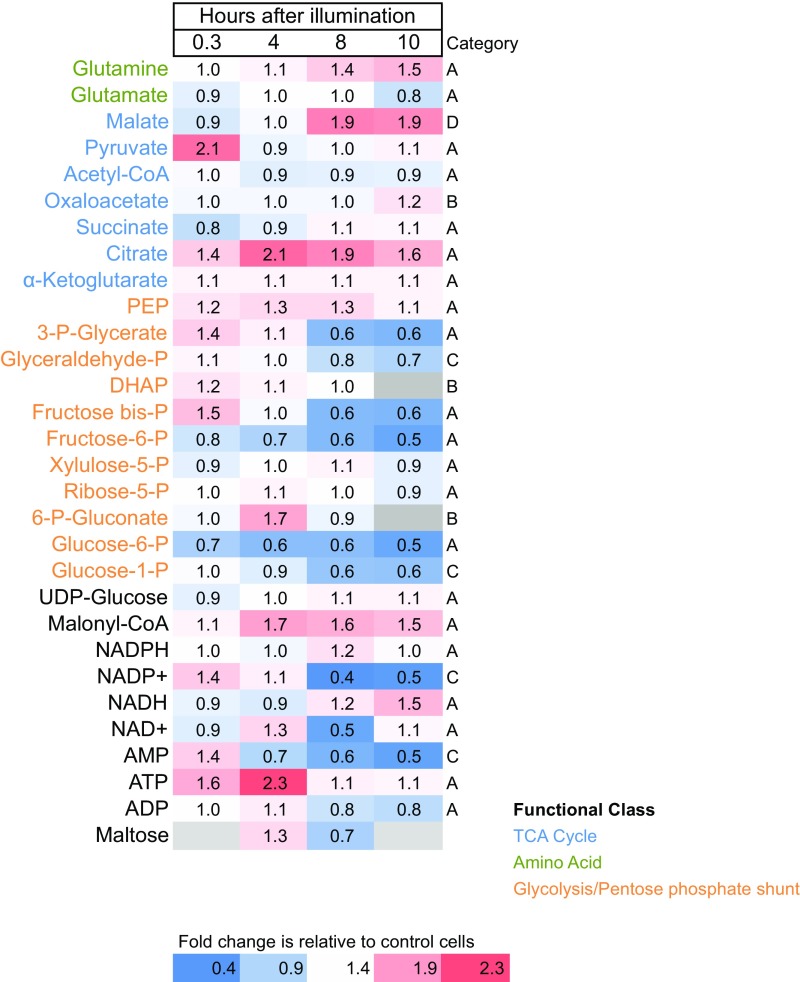
To understand how cell cycle arrest affects intermediate metabolism, we analyzed metabolite pools on returning synchronized (dark-arrested in G1) cells to light. The largest differences in metabolites between control and NU 2058-treated cells were associated with the pentose phosphate pathway, glycolysis, and the tricarboxylic acid (TCA) cycle (Fig. 6). The metabolites most affected were pyruvate, phosphoenolpyruvate, 3-phosphoglycerate, and fructose bis-phosphate, suggesting an initial buildup of recently fixed photosynthate through carbon-fixation and/or glycolysis. Only five metabolites were found at higher levels in treated cells compared with controls: malate, citrate, glutamine, malonyl-CoA, and NADH. Glutamine, the first intermediate in the assimilation of nitrogen in diatoms (19), was 50% higher in treated cells after 10 h of exposure to the inhibitor. Malonyl-CoA, which provides two-carbon units during fatty acid elongation, was approximately 50% higher in treated cells at 10 h; however, acetyl-CoA levels were the same or slightly lower than those in untreated cells.
Fig. 6.
Fold change in metabolites. Synchronized cells were exposed to continuous light and sampled at several intervals for metabolites, in both untreated control and 7 µM NU 2058-treated cells. Category A: metabolites that rise after illumination in both control and treated cells. Category B: metabolites that fall with illumination in both control and treated cells. Category C: metabolites that rise in control cells and fall or stay the same in treated cells. Category D: metabolites that rise in treated cells and fall in control cells.
Other large differences were in AMP, which was 50% lower in treated cells compared with untreated cells. NADP+ levels in treated cells were approximately one-half those in control cells, and NADH levels were roughly 50% higher in treated cells. Most metabolites increased with illumination in both control and treated cells (category A). Three metabolites—oxaloacetate, dihydroxyacetone-phosphate, and 6-phospho-gluconate—decreased with illumination (category B). In this category, there was no large difference between control and treated cells. Only one measured metabolite—malate—tended to rise in treated cells and fall in control cells (category D). Of the glycolysis/pentose phosphate metabolites, five—phosphoenolpyruvate, 3-phosphoglycerate, glyceraldehyde-3-phosphate, fructose-bis-phosphate, and dihydroxyacetone-phosphate—increased initially in arrested cells, but decreased gradually over time.
Transcriptome.
To better understand the effect of the cell cycle on gene expression, cells were synchronized in G1 phase by prolonged darkness (24 h) and then separated into cultures with or without NU 2058. At the time of release from G1 phase by illumination (t0), cells (100% in G1 phase) were collected for transcriptome and cell cycle analysis (Dataset S1). After 4 h of illumination, cells were again collected for mRNA and cell cycle analysis. The cells illuminated for 4 h were designated “t4” for cultures without the inhibitor and “N” for cultures with NU 2058. At this time point, t4 cells were roughly 79% in G1 phase and 20% in S phase, and N cells were 95% in G1 phase and 5% in S phase. In addition, a culture with the inhibitor was maintained in parallel to obtain mRNA for arrested cells at 20 h after illumination (“ND”). This culture had 70% of cells in G1 phase, 18% in S phase, and 12% in G2 phase.
Cell Cycle Genes.
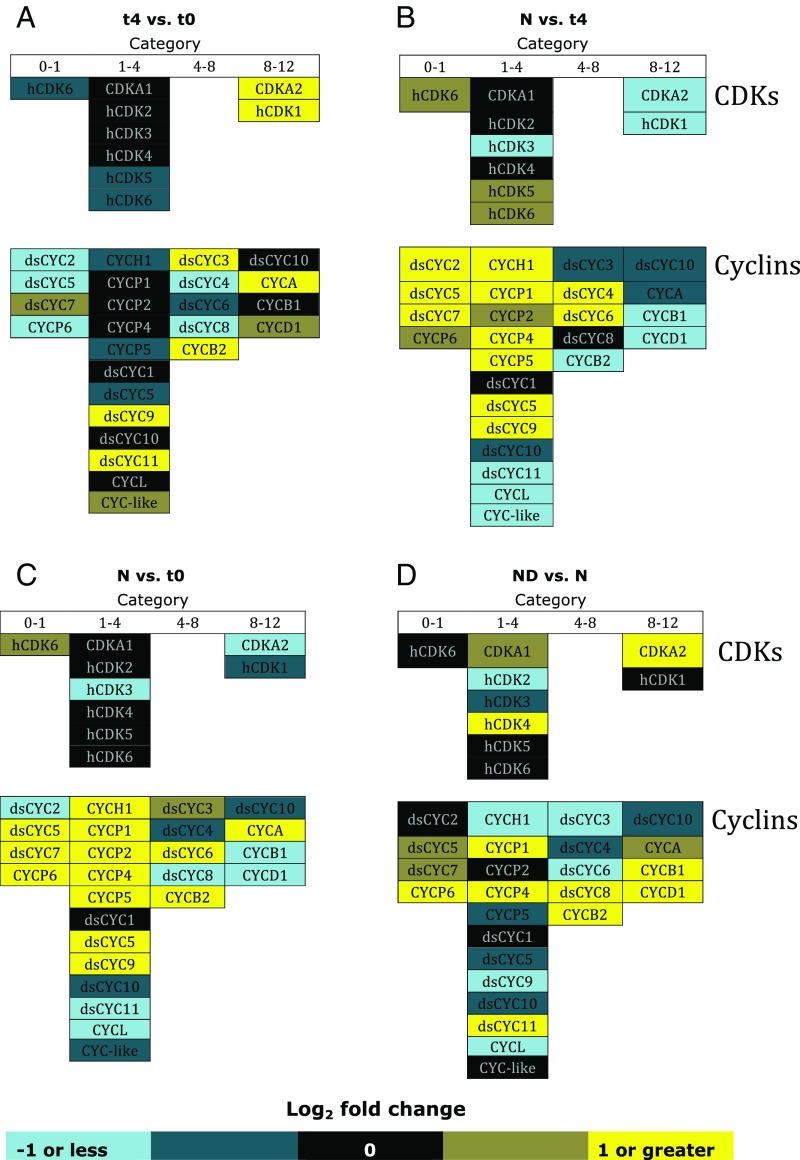
Cell cycle genes were categorized into four groups based on the timing of maximal accumulation of transcripts (16): (i) the time immediately following the onset of light (early G1), 0–1 h after illumination; (ii) G1, 1–4 h after illumination; (iii) late G1 and S phase, 4–8 h after illumination; and (iv) G2/M, 8–12 h after illumination. The general pattern of transcription for t4 cells showed that genes in the first category were down-regulated and genes of category 4 were up-regulated compared with cells at t0 (Fig. 7A). This result might be expected given that t0 cells would be relatively enriched for category 1 transcripts, and t4 cells should be relatively enriched for later category transcripts. However, two category 3 genes (dsCYC4 and dsCYC8, which are diatom-specific cyclins) were not consistent with this general trend, and were down-regulated at t4 compared with t0.
Fig. 7.
Regulation of cell cycle genes. Cell cycle genes were categorized by time of occurrence of peak transcription in synchronized cells and the corresponding phase of the cell cycle, as follows: early G1 (0–1 h after illumination); G1 (1–4 h after illumination); late G1/S phase (4–8 h after illumination); and G2/M (8–12 h after illumination). These categories were based on the P. tricornutum CDK and cyclin expression profiles of Huysman et al. (16). Transcript level comparisons: (A) control 4 h vs. t0; (B) NU 2058-treated 4 h vs. control 4 h; (C) NU 2058-treated 4 h vs. t0; and (D) NU 2058-treated 20 h vs. 4 h. ds, diatom specific; h, hypothetical.
At 4 h after the return of cultures to the light, many of the cyclin genes were expressed at higher levels in the NU2058-treated cells relative to t4 control cells (Fig. 7B).
CDKA2 and hCDK1 are thought to play a role in G2 and M phases because transcripts for these two CDKs normally peak between 8 and 12 h after illumination (16). Moreover, CDKA2 plays a role in mitosis, specifically in forming the cleavage furrow or in recruiting components for the cell division plane, and its overexpression has been found to prolong mitosis (17). Since ND cells are a later time point compared with N cells, a greater number of later category genes were up-regulated, including CDKA2 (Fig. 7D).
Expression of Light-Harvesting Complex and Reactive Oxygen Species Scavenging Genes.
P. tricornutum is predicted to have at least 40 genes that encode for light-harvesting complexes (LHCs). These include the LHCFs, encoding the major fucoxanthin Chl a/c proteins, the red algal-like LHCRs, and the LI818-like LHCXs. The majority of LHCs in t4 cells (roughly 76% of LHC genes) were up-regulated compared with cells at t0. In N cells, however, close to 80% of LHC genes were down-regulated compared with t4 cells, even though LHCR8 and LHCX3 were up-regulated. The general pattern of LHC expression is similar to that seen in cells stressed by high light (20).
Of the genes involved in reactive oxygen species (ROS) scavenging, approximately 67% were up-regulated and 22% were down-regulated in N cells compared with t0 cells, and 67% were up-regulated and 25% were down-regulated in N cells compared with t4 cells (Dataset S1:3), indicating that N cells were under greater oxidative stress than t0 or t4 cells.
Gene Expression of Calvin Cycle, Pentose Phosphate, Lipid, and Chrysolaminarin Pathways.
Compared with t4 cells, N cells down-regulated most genes of the Calvin cycle, including phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, triosephosphate isomerase, and fructose 1,6-bisphosphate aldolase, by more than threefold (Dataset S1:10). N cells also down-regulated most genes for the pentose phosphate pathway in the cytosol, compared with both t0 and t4 cells (Dataset S1:14). This finding is consistent with metabolite profiles showing that immediately after the onset of light, there was a buildup of most pentose phosphates in N cells compared with control cells, but with increasing time in the light, levels of these metabolites were lower than levels in control cells.
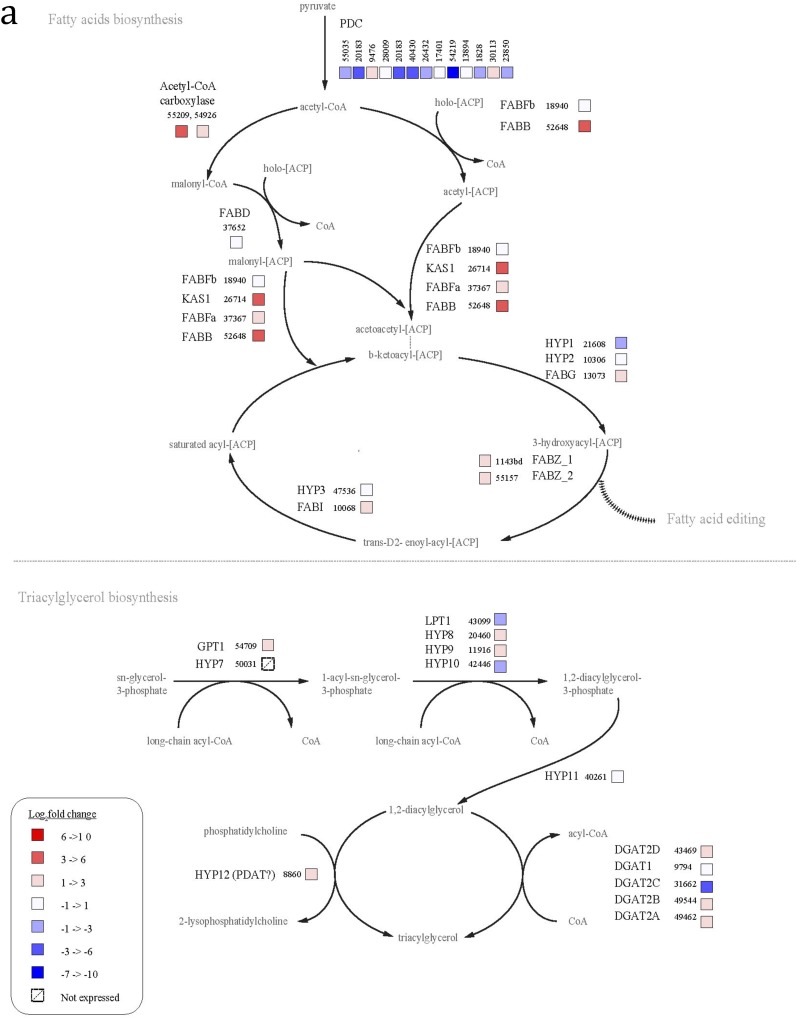
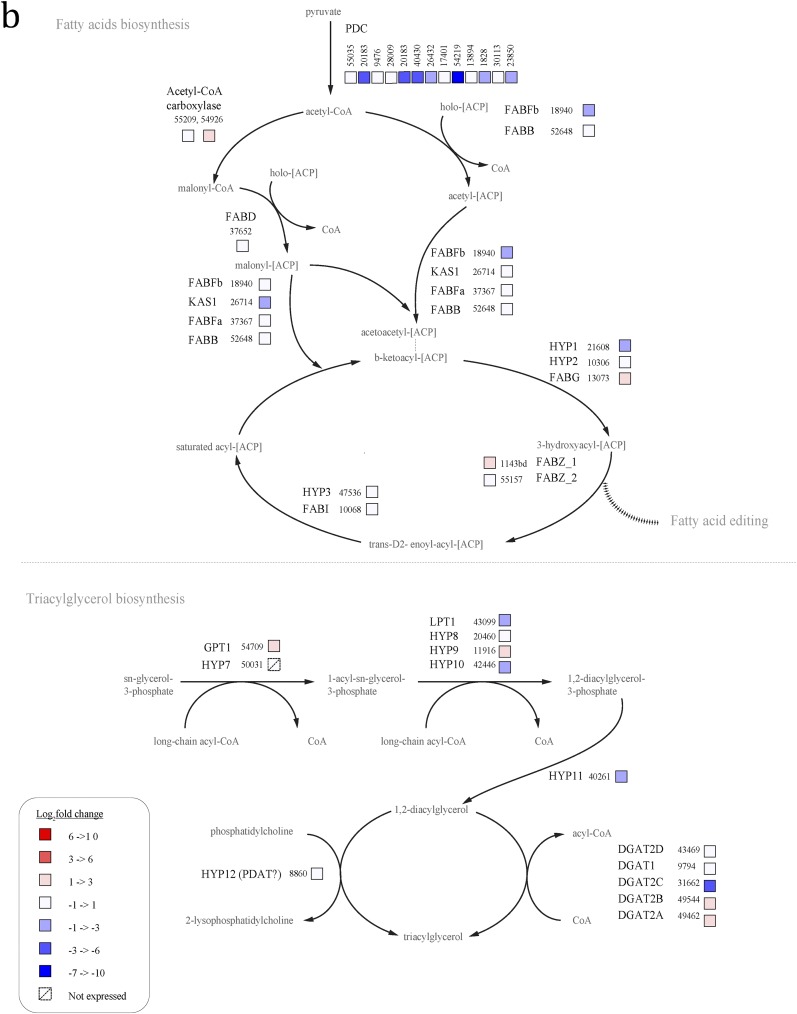
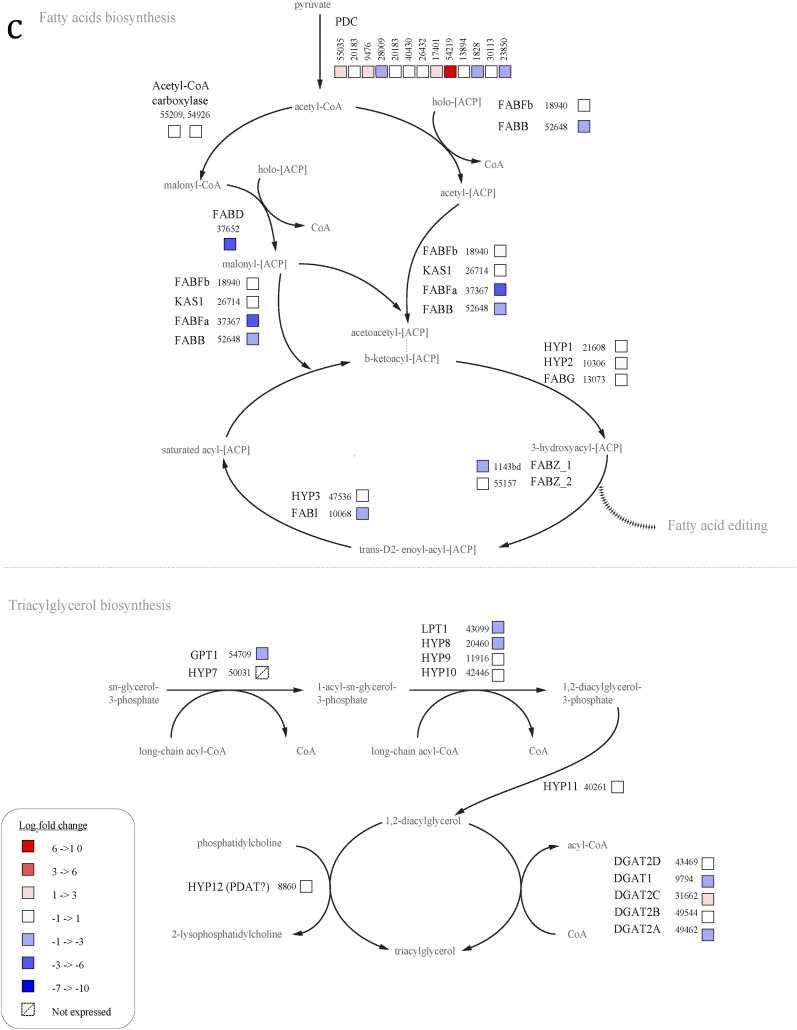
N cells up-regulate most genes involved in fatty acid synthesis, compared with t4 cells (Fig. S1A). The genes involved in TAG metabolism are not well annotated, but of the five known diacylglycerol acyltransferases (DGATs) in P. tricornutum, four were up-regulated in N cells (43469, 49462, 49544, and 9794) and one was down-regulated (31662), compared with t4 cells. Acetyl-CoA carboxylase (55209 and 54926), which performs the first committed step in fatty acid synthesis, was up-regulated as well.
Fig. S1.
Differential expression of lipid synthesis genes N vs. t4 (A), N vs. t0 (B), and ND vs. N (C). Figure adapted from ref. 12.
In contrast, most genes involved in fatty acid synthesis were not differentially expressed in N cells relative to t0 cells, and some were even down-regulated, indicating that many of these fatty acid synthesis genes are highly up-regulated in untreated cells immediately at the onset of light (Fig. S1B). Many of the lipid synthesis genes up-regulated in N cells were down-regulated or not differentially expressed in ND cells (Fig. S1C). These include ACC1 (54926), ACC2 (55209), four DGATs (43469, 49462, 49544, and 9794), and most of the genes for fatty acid synthesis.
Integration of Gene Expression Data with the Flux Balance Analysis Model.
To better understand the effects of cell cycle arrest on the fluxes of intermediate metabolites, we used a flux balance analysis based on a model previously developed for P. tricornutum (21). The predicted fluxes from E-Flux2 are listed in Dataset S2:1. Biomass equations were adjusted using experimental data (Dataset S2:2). The biomass equation for t0 was set as the “average” cell as formulated for the autotrophic cell. The t4 biomass equation was formulated relative to t0 cells; that is, the “goal” was to model the fluxes that produce the biomass of a cell at t0, minus the macromolecular components (DNA, amino acids, lipids, etc.) that were synthesized between t0 and t4. For the N and ND conditions, the differences in macromolecules between the cells at t0 and the measured macromolecules at the respective time points for N and ND were taken to be the goal and were designated version 1 of N and version 1 of ND, respectively. Since the true “biological objectives” for N and ND conditions were unknown, a sensitivity analysis was performed to compare several simulations of the model using different biomass equations for these conditions.
The sensitivity analysis revealed that the differences in these versions did not change the overall flux pattern, but did change the rate of flux through reactions (Dataset S2:3). Changing the ATP growth requirement did not change the biomass output, because when photon and carbon input fluxes are left unconstrained, any increased ATP demands could be met by a combination of increased photon and/or carbon input to increase linear and/or cyclic electron flow. This model would be expected to reliably predict the overall flux distribution pattern, but not to predict the specific quantitative values of individual fluxes.
Overall Flux Predictions.
Although the overall flux distributions (Dataset S2) were similar to those predicted previously (21), there were differences in some of reactions, especially those involving the mitochondria. This organelle is a source of glycerate that ultimately goes into the production of glycerol-3-phosphate, which can be used to synthesize polar or neutral lipids. The model predicts that serine-pyruvate transaminase and glycerate dehydrogenase in the mitochondrion support TAG synthesis by providing a source of glycerate. Moreover, whenever there is flux through serine-pyruvate transaminase, glycerate dehydrogenase, and/or glycerate kinase in the mitochondria, the enzymes of glycolysis in the mitochondrion run in the direction of gluconeogenesis to export triose phosphates. There is no other outlet for the glycerate-3-phosphate produced in the mitochondrion. Gluconeogenesis also results in the conversion of pyruvate and serine into alanine and hydroxypyruvate, and supports mitochondrial electron transport and the net removal of protons from the inner mitochondrial matrix.
Discussion
Under nutrient-replete conditions in the light, unicellular photoautotrophic algae progress through the cell cycle, leading to cell replication and balanced growth. Balanced growth is achieved when cells reach a steady-state biochemical composition; that is, the rate of growth of any cell constituent, such as protein, lipid, or DNA, remains the same if averaged over a generation or more (22). Specific phases of the cell cycle are used to synthesize specific macromolecular pools. The G1 phase is critical for protein biosynthesis. Normally, cells in G1 direct a significant portion of carbon in intermediate metabolites to pyruvate, which is decarboxylated to form AcCoA. This is a key branch point; the AcCoA can enter the TCA cycle to form intermediate metabolites for both anabolic and anaplerotic pathways, especially in amino acid biosynthesis, or it can be directed toward lipid biosynthesis (23). Under optimal growth conditions, carbon is preferentially allocated to proteins.
The results presented here reveal that when a diatom cell is arrested in G1 phase, photosynthetically generated products are preferentially shunted into neutral lipids and chrysolaminarin. Arrested cells have lower electron transport rates and higher nonphotochemical quenching, suggesting that they have a smaller sink for electrons compared with actively dividing cells. Indeed, arrested cells have lower protein and nucleotide pools compared with untreated cells that progress through the cell cycle.
This study demonstrates that cell cycle arrest in G1 is sufficient to alter the fate of intermediate metabolites, leading to increased lipid content without significantly affecting maximal photosynthetic energy conversion efficiency. This phenomenon is reminiscent of strains with knocked-down nitrate reductase activity (12). Indeed, arrested cells have lower NR activity and lower nitrate assimilation rates. The reduction in NR activity is potentially a feedback with lipid accumulation; changes in NR activity and cellular carbon content are strongly negatively correlated in the diatom Thalassiosira weissflogii with a time lag of 6 h (24). However, unlike cells arrested in G1, NR knockdown strains ultimately achieve balanced growth. In cells arrested in G1, the ratio of fatty acid content in arrested to control cells is 1.8-fold higher than that in NR21 to WT cells, suggesting that cell cycle arrest is more disruptive to intermediate metabolism than reduced nitrogen assimilation.
A key branch point in lipid biosynthesis in P. tricornutum is in GS/GOGAT/GDH (glutamine synthetase/glutamine oxoglutarate aminotransferase/glutamate dehydrogenase) (10, 12). Glutamine is the first organic nitrogen carrier in diatoms (19), and its utilization leads to the formation of other amino acids. Under nitrogen stress or NR knockdown, P. tricornutum increases its GLU/GLN ratios by 12-fold or more (10, 12, 25, 26). In general, a higher GLU/GLN ratio has consistently been associated with N deprivation in a number of different algae, and has been proposed as a biomarker for N-depleted cells (27). In contrast, cells treated with NU 2058 have a 41% lower GLU/GLN ratio compared with control cells. Thus, the GLU/GLN ratio shows opposite tendencies in N-stressed and N-arrested cells. This may have to do with the fact that N-stressed cells have a higher catabolism of enzymes that are part of the photosynthetic machinery, releasing intracellular nitrogen that can be redirected into enzymes that can confer a selective advantage under nitrogen stress (12). More generally, the GLU/GLN ratio may be not just a biomarker for N stress, but also an indicator of how much nitrogen is available relative to the nitrogen demand.
In conclusion, our experimental results show that when diatom cells are arrested in G1 phase, they accumulate storage lipids (TAGs), up-regulate most genes involved in fatty acid synthesis, build up pentose phosphates immediately upon the onset of light, have lower protein and nitrate reductase activity, and down-regulate most genes involved in nitrogen metabolism. Although cell cycle arrest does not lead to changes in key photosynthetic attributes, it does lead to lower electron transport rates, higher nonphotochemical quenching, and LHC/ROS scavenging gene expression patterns similar to cells stressed by high light, suggesting that arrested cells have a smaller sink for photosynthetically generated electrons compared with actively growing cells. This is in contrast to nitrogen-starved cells, which cannibalize their photosynthetic apparatus and accumulate lipids via carbon reallocation.
This study shows that cell cycle arrest alone is sufficient to increase TAG accumulation in a diatom. Hence, unlike in higher plants, while the overall organism can often grow under an array of conditions, unbalanced production of a macromolecular source can often be transported and stored in specific structures or organs that act as sinks. In a unicellular alga, such as a diatom, the sources and sinks are tightly coupled to the cell cycle, and disruption of the latter inevitably leads to unbalanced growth. Our results clearly show that in the specific case of a diatom, cell cycle arrest in a G phase does not significantly alter photosynthetic energy conversion efficiency, but does lead to a remodeling of intermediate metabolism that results in the accumulation of intracellular storage of lipids.
Materials and Methods
Experimental Conditions.
P. tricornutum, strain CCAP 1055/1, was grown axenically in filtered and autoclaved seawater supplemented with F/2 medium (28). Cultures were aerated through 0.2-µm filters and maintained at 18 °C under continuous light (∼90 µmol photons m2 s−1). All purine analogs were administered with 0.1% DMSO vol/vol to facilitate dissolution in seawater. Control cultures also had a final DMSO concentration of 0.1%.
Analytical Methods.
Growth rates were determined by cell counts measured with a Beckman Coulter Multisizer 3. For experiments requiring synchronized cultures, cells were arrested in G1 phase by prolonged darkness (24 h) (16). On return of cultures to light, inhibitor (NU 2058) was added to test cultures (0.1% DMSO for control cultures). This time point was designated t0. Synchronization was validated by flow cytometry cell cycle analysis with an Influx Mariner 209S flow cytometer (BD Biosciences) equipped with a 488-nm laser, in cells fixed in methanol, washed three times with 1× PBS, treated with RNase A for 1 h, and stained with propidium iodide. For each sample, roughly 10,000 cells were processed. Gating and data analyses were performed using FlowJo analytical software.
Cellular carbon and nitrogen content were measured on cells collected onto precombusted 13-mm A/E filters (Pall Gelman) and analyzed on a CHN analyzer (NA 1500 series 2; Carlo Erba Instruments). Total FAMEs were measured by collecting 5 × 107 cells onto 25-mm Whatman GF/F filters and extracting them in 10-mL glass vials with 2 mL of 1:20 (vol:vol) acetyl chloride-methanol mixture and 1 mL of hexane in the presence of 25 µg of heptadecanoic acid (Sigma-Aldrich), which served as an internal standard (29). The vials were boiled in a water bath for 1 h. After cooling, 1 mL of MilliQ water was added, and the hexane phases were transferred into gas chromatograph vials with Teflon-lined caps (Thermo Fisher Scientific). Fatty acid analysis was performed by gas chromatography [GC 2010; Shimadzu; equipped with a TR-FAME (0.25 µm × 60 m) column (Thermo Fisher Scientific) and a flame ionization detector]. Fatty acids were identified by reference to verified standards (Supelco 37 component FAME; Sigma-Aldrich) processed and analyzed under the same conditions. TAGs were separated using thin-layer chromatography (30), with modifications as described by Guerra et al. (10). IPLs were separated using stepwise elutions (31). All lipids were compared with commercial standards from Sigma-Aldrich and Avanti Polar Lipids.
Carbohydrate quantification was based on a modified phenol sulfuric acid assay (32). In brief, 107 cells were filtered onto polycarbonate filters and sonicated three times for 20 s each time in 300 µL of MilliQ water while kept on ice. For each sample, 10 µL of cell lysate was transferred to a microplate with 2 µL of 90% phenol solution, 70 µL of MilliQ water, and 200 µL of sulfuric acid. Plates were incubated for 30 min at room temperature, and the absorbance was read at 485 nm and compared with glucose standards.
For protein quantification, 1 × 108 cells were filtered onto polycarbonate filters, and protein was extracted with 300 µL of extraction buffer (2% lithium dodecyl sulfate, 100 mM Tris base, 105 mM Tris⋅HCl, 0.5 mM EDTA, 10% glycerol, and 15 µL of protease inhibitor mixture solution; Sigma-Aldrich). Cell suspensions were sonicated three times in between liquid nitrogen freezes (Microson ultrasonic cell disruptor, set at power level 2), then centrifuged at 16,000 × g for 5 min. Protein concentrations were determined with the Bio-Rad DC Protein Assay. For immunoblots, protein extracts were heated for 5 min with 30 µL of 1 M DTT and 300 µL of a solution containing of 4% SDS, 15% glycerol, and 0.05% bromothymol blue. Protein (2 µg) was loaded onto a precast 4–20% Tris⋅HCl gel (Bio-Rad), run for 1 h (100 mV), and then transferred electrophoretically to a PVDF membrane. Antibodies toward PsbD and RbcL (Agrisera AS06 146 and AS03 037, respectively) were used at a 1:20,000 dilution. PSTAIR antibody (Sigma-Aldrich; P7962) was used at 1:4,000.
NR activity was determined colorimetrically and calculated as µmol NO2− produced per minute per cell as described by Berges and Harrison (33). NU 2058 was added to control cells immediately before harvesting for NR activity assays (at a final concentration of 7 µM), to determine whether NU 2058 has a direct effect on NR enzyme activity.
The maximum photochemical quantum yields of PSII (Fv/Fm) were measured on dark-adapted cell suspensions with single turnover saturating flashes using a fluorescence induction and relaxation system (Satlantic) (34). For chlorophyll measurements, 10–20 mL of culture was collected by vacuum filtration onto Whatman 25-mm GF/F filters. Chlorophyll was extracted in 90% acetone and measured spectrophotometrically following the equations of Jeffrey and Humphrey (35). The in vivo absorption spectra were measured with an SLM-Aminco DW-2000 spectrophotometer, using live cell suspensions. The values obtained were used to calculate the wavelength-specific optical absorption cross-section normalized to Chl a (a*) (36, 37).
For affinity chromatography, protein extracts were loaded over NU 2058 immobilized to CNBr-activated sepahrose matrix and then sequenced. The extracts contained protein from a mixture of cells in G1, S, and G2 phases (roughly 40%, 25%, and 35%, respectively). NU 2058 resins were prepared as described by Bach et al. (38). For affinity chromatography, 1.5 mg of protein extracts were used for 15 µL of packed beads. Protein kinase assays were performed as described previously (38).
Metabolite profiles were obtained as described by Bennette et al. (39). Cells were quenched via rapid filtration of 15 mL of cells, followed by immediate transfer of filters to 1.8 mL of 80:20 (vol:vol) methanol:water at −20 °C in 35 × 10-mm Petri dishes. Filters were incubated at −20 °C for 15 min and then scraped clean of cells. Then 1.8 mL of extraction solvent and cells was transferred to 2-mL Eppendorf tubes, followed by a washing of the filter with 0.25 mL of 80:20 methanol:water at −20 °C to collect remaining cell material. The tubes were spun at 4 °C, and the supernatant was collected. To the cell pellet, 100 µL of 80:20 methanol:water at −20 °C was added, followed by vortexing, incubation at −20 °C for 15 min, and pelleting. This 100 µL was combined with the original supernatant, evaporated in a SpeedVac, and resuspended in 33 µL of water, which was transferred to gas chromatograph vials.
Analysis of the metabolites was done in one 35-min LC-MS/MS run on an Agilent 1200 series liquid chromatograph with a 6410 QQQ mass spectrometer using reversed-phase ion-pairing chromatography. Standards for each metabolite, run with sample background, were used for quantification. The column used was a Synergi 2.5-µm hydro RP 100A, size 100 × 2 mm (Phenomonex).
Calculation of the Quantum Requirement for Carbon Allocation into Lipids.
The efficiency of transduction of light energy at a given irradiance to fatty acid was calculated with a modified version of the model proposed by Falkowski et al. (37), and expressed as the quantum requirement for fatty acid biosynthesis, 1/ϕFA:
| [1] |
where ϕFA is the quantum yield for fatty acid biosynthesis (mol C−1), a* is the in vivo absorption cross-section (m2 mg−1 Chl a), Chl a and C represent the biomass (mg) of Chl a and carbon [we calculated the C stored as fatty acid, assuming oleic acid (C18:1; molecular weight 282,46 g mol−1) as the average fatty acid molecule], Iµ is the growth irradiance (µmol quanta m−2 s−1), μ is the specific growth rate (d−1), and the constant 1,040 was used to convert units from μmol quanta m−2 s−1 to mol quanta m−2 d−1 and from mg C to mol C. Higher 1/ϕFA values indicate decreased efficiency of light energy transduction into fatty acid production, and lower values indicate increased efficiency.
RNA-Seq.
P. tricornutum cultures synchronized in G1 phase (subjected to darkness for 24 h) were divided into cultures with and without NU 2058, placed in the light (∼90 µmol photons m2 s−1), and collected for RNA. Samples for RNA-Seq from two independent biological replicates of each treatment were harvested by centrifuging cultures at 8,000 × g using a Sorvall centrifuge at 4 °C, then transferring them to RNase and DNase-free microtubes that were immediately flash-frozen in liquid N2. RNA was extracted using RLT lysis buffer and the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions, followed by removal of DNA contaminants with Ambion Turbo DNase (AM1907; Life Technologies). The TruSeq RNA Sample Preparation Kit 2 (Illumina) was used to prepare mRNA libraries for each of the samples according to the manufacturer’s instructions. The 50-bp single-ended libraries were multiplexed and sequenced on an Illumina HiSeq2000 platform. The raw reads were trimmed for low-quality and adaptor sequences and aligned to P. tricornutum’s version 2.0 set of 10,402 filtered gene models using CLC Genomics Workbench v6.02 (40). Files were filtered to include uniquely aligned reads with no more than three mismatches. Gene counts (unique aligned reads per gene) were used for differentially expressed (DE) analysis carried out using the DESeq R/Bioconductor package (40), which infers DE based on the negative binomial distribution. A cutoff of 5% was used to control for false detection rate (false positives), and only genes that had a log twofold change ≥ ±1 were considered DE.
Computation of Metabolic Fluxes.
The P. tricornutum flux balance model (21) was correlated with transcriptome data using E-Flux2, as described by Kim et al. (41). In brief, with E-Flux2, mRNA levels served as upper bounds on the maximum amount of metabolic enzymes (reaction rates). The calculations performed had numerical tolerances such that there was a round-off error associated with all values. The lower bound on the uncertainty was 10−9; thus, all flux values had an uncertainty of at least 10−9 plus the uncertainties associated with the transcriptome measurements. Comparisons across conditions were made by first normalizing all fluxes to the flux of total incoming inorganic carbon.
In total, two versions of N and four versions of ND were tested. Version 2 for N and ND was the same as version 1, except that the growth-associated ATP maintenance, the unknown ATP requirements associated with biosynthesis, polymerization, and transport, was one-third of the original in version 1, which was set to 29.89 mmol ATP/g dry wt/h based on the work of Kim et al. (21). Version 2 of N and ND was established to analyze flux outputs when ATP requirements were drastically lower, since it is likely that growth arrest would lower growth-associated ATP maintenance. The other two versions for ND (versions 3 and 4) were the same as versions 1 and 2 for ND, respectively, except that the biomass was set to be the difference in macromolecules between the cell at N (instead of t0) and the measured biomass at the ND time point.
Supplementary Material
Acknowledgments
We thank Miguel J. Frada, Daniel Vaulot, Olivier Lozach, Orly Levitan, Udi Zelzion, Benjamin Bailleul, Xiao Qian, Marie Huysman, Lieven De Veylder, and James Kelly for constructive discussions and technical support, and Raymond M. Remus for synthesis of NU 2058. We also thank the Cancéropôle Grand-Ouest (axis: natural sea products in cancer treatment), les Groupements d’Intérêt Scientifique Infrastructures en Biologie Santé et Agronomie, and Biogenouest for supporting the KISSf screening facility (Roscoff, France). S.B. is supported by the ANR/Investissements d’Avenir program through the OCEANOMICs project (Grant ANR-11-BTBR-0008). This research was supported by Rutgers-National Science Foundation Integrative Graduate Education and Research Traineeship Project NSF DGE O903675 (Eric Lam, PI; P.G.F., co-PI) and by the Bennett L. Smith Endowment (to P.G.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711642114/-/DCSupplemental.
References
- 1.Wiebe HCF, Kooistra RG, Medlin LK, Mann DG. In: Evolution of Primary Producers in the Sea. Falkowski PG, Knoll AH, editors. Elsevier; Amsterdam: 2007. pp. 207–249. [Google Scholar]
- 2.Thomas WH, Dodson AN, Reid FMH. Diatom productivity compared to other algae in natural phytoplankton assemblages. J Phycol. 1978;14:250–253. [Google Scholar]
- 3.Margalef R. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanol Acta. 1978;1:493–509. [Google Scholar]
- 4.Parsons TR, Stephens K, Strickland JDH. On the chemical composition of eleven species of marine phytoplankters. J Fish Res Bd Can. 1961;18:1001–1016. [Google Scholar]
- 5.Badour SS, Gergis MS. Cell division and fat accumulation in Nitzschia sp. grown in continuously illuminated mass cultures. Arch Mikrobiol. 1965;51:94–102. doi: 10.1007/BF00406853. [DOI] [PubMed] [Google Scholar]
- 6.Fogg GE. Photosynthesis and formation of fats in a diatom. Ann Bot. 1956;20:265–285. [Google Scholar]
- 7.Opute FI. Studies on fat accumulation in Nitzschia palea Kutz. Ann Bot. 1974;38:889–902. [Google Scholar]
- 8.Shifrin NS, Chisholm SW. Phytoplankton lipids: Interspecific differences and effects of nitrate, silicate and light-dark cycles. J Phycol. 1981;17:374–384. [Google Scholar]
- 9.Lynn SG, Kilham SS, Kreeger DA, Interlandi SJ. Effect of nutrient availability on the biochemical and elemental stoichiometry in the freshwater diatom Stephanodiscus minutulus (Bacillariophyceae) J Phycol. 2000;36:510–522. doi: 10.1046/j.1529-8817.2000.98251.x. [DOI] [PubMed] [Google Scholar]
- 10.Guerra LT, et al. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy. 2013;59:306–315. [Google Scholar]
- 11.Sheehan J, Dunahay T, Benemann J, Roessler P. 1998. A look back at the US Department of Energy’s Aquatic Species Program: Biodiesel from algae. Report NREL/TP-580-24190 (National Renewable Energy Laboratory, Golden, CO)
- 12.Levitan O, Dinamarca J, Zelzion E, Gorbunov M, Falkowski PG. An RNAi knock-down of nitrate reductase enhances lipid biosynthesis in the diatom Phaeodactylum tricornutum. Plant J. 2015;84:963–973. doi: 10.1111/tpj.13052. [DOI] [PubMed] [Google Scholar]
- 13.King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 14.Arris CE, et al. Identification of novel purine and pyrimidine cyclin-dependent kinase inhibitors with distinct molecular interactions and tumor cell growth inhibition profiles. J Med Chem. 2000;43:2797–2804. doi: 10.1021/jm990628o. [DOI] [PubMed] [Google Scholar]
- 15.Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 16.Huysman MJJ, et al. Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biol. 2010;11:R17. doi: 10.1186/gb-2010-11-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huysman MJJ, Tanaka A, Bowler C, Vyverman W, De Veylder L. Functional characterization of the diatom cyclin-dependent kinase A2 as a mitotic regulator reveals plant-like properties in a non-green lineage. BMC Plant Biol. 2015;15:86. doi: 10.1186/s12870-015-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levitan O, et al. Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc Natl Acad Sci USA. 2015;112:412–417. doi: 10.1073/pnas.1419818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zehr JP, Capone DG, Falkowski PG. Rapid incorporation of 13NO3 by NH4-limited phytoplankton. Mar Ecol Prog Ser. 1989;51:237–241. [Google Scholar]
- 20.Nymark M, et al. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS One. 2009;4:e7743. doi: 10.1371/journal.pone.0007743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, et al. Flux balance analysis of primary metabolism in the diatom Phaeodactylum tricornutum. Plant J. 2016;85:161–176. doi: 10.1111/tpj.13081. [DOI] [PubMed] [Google Scholar]
- 22.Eppley RW. Relations between nutrient assimilation and growth in phytoplankton, with a brief review of estimates of growth rate in the ocean. Can Bull Fish Aquat Sci. 1981;210:251–263. [Google Scholar]
- 23.Turpin DH, Dennis DT. Plant Physiology, Biochemistry and Molecular Biology. Longman Group; London: 1991. [Google Scholar]
- 24.Vergara JJ, Berges JA, Falkowski PG. Diel periodicity of nitrate reductase activity and protein levels in the marine diatom Thalassiosira weissflogii (Bacillariophyceae) J Phycol. 1998;34:952–961. [Google Scholar]
- 25.Flynn KJ, Al-Amoudi OA. Effects of N deprivation and darkness on composition of free amino acid pool in and on amino acid release from diatom Phaeodactylum tricornutum Bohlin. J Exp Mar Biol Ecol. 1988;119:131–143. [Google Scholar]
- 26.Frada MJ, Burrows EH, Wyman KD, Falkowski PG. Quantum requirements for growth and fatty acid biosynthesis in the marine diatom Phaeodactylum tricornutum (Bacillariophyceae) in nitrogen-replete and limited conditions. J Phycol. 2013;49:381–388. doi: 10.1111/jpy.12046. [DOI] [PubMed] [Google Scholar]
- 27.Flynn KJ, Dickson DMJ, Al-Amoudi OA. The ratio of glutamine:glutamate in microalgae: A biomarker for N status suitable for use at natural cell densities. J Plankton Res. 1989;11:165–170. [Google Scholar]
- 28.Guillard RR, Ryther JH. Studies of marine planktonic diatoms, I: Cyclotella nana Hustedt and Detonula confervacea (cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Ruiz J, Belarbi EH, Sanchez JLG, Alonso DL. Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol Tech. 1998;12:689–691. [Google Scholar]
- 30.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yongmanitchai W, Ward OP. Separation of lipid classes from Phaeodactylum tricornutum using silica cartridges. Phytochemistry. 1992;31:3405–3408. [Google Scholar]
- 32.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 33.Berges J, Harrison PJ. Nitrate reductase activity quantitatively predicts the rate of nitrate incorporation under steady state light limitation: A revised assay and characterization of the enzyme in three species of marine phytoplankton. Limnol Oceanogr. 1995;40:82–93. [Google Scholar]
- 34.Gorbunov MY, Falkowski PG. Fluorescence induction and relaxation (FIRe) technique and instrumentation for monitoring photosynthetic processes and primary production in aquatic ecosystems. In: van der Est A, Bruce D, editors. Photosynthesis: Fundamental Aspects to Global Perspectives: Proceedings of the 13th International Congress of Photosynthesis. Allen Press; Oxford, UK: 2004. [Google Scholar]
- 35.Jeffrey SW, Humphrey GF. New spectrophotometric equations for determining chlorophylls a, b, c1, and c2 in higher plants, algae, and natural phytoplankton. Biochem Physiol Pflanz. 1975;167:191–194. [Google Scholar]
- 36.Dubinsky Z, Falkowski PG, Wyman K. Light harvesting and utilization by phytoplankton. Plant Cell Physiol. 1986;27:1335–1349. [Google Scholar]
- 37.Falkowski PG, Dubinsky Z, Wyman K. Growth-irradiance relationships in phytoplankton. Limnol Oceanogr. 1985;30:311–321. [Google Scholar]
- 38.Bach S, et al. Roscovitine targets, protein kinases and pyridoxal kinase. J Biol Chem. 2005;280:31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 39.Bennette NB, Eng JF, Dismukes GC. An LC-MS-based chemical and analytical method for targeted metabolite quantification in the model cyanobacterium Synechococcus sp. PCC 7002. Anal Chem. 2011;83:3808–3816. doi: 10.1021/ac200108a. [DOI] [PubMed] [Google Scholar]
- 40.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MK, Lane A, Kelley JJ, Lun DS. E-Flux2 and SPOT: Validated methods for inferring intracellular metabolic flux distributions from transcriptomic data. PLoS One. 2016;11:e0157101. doi: 10.1371/journal.pone.0157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.