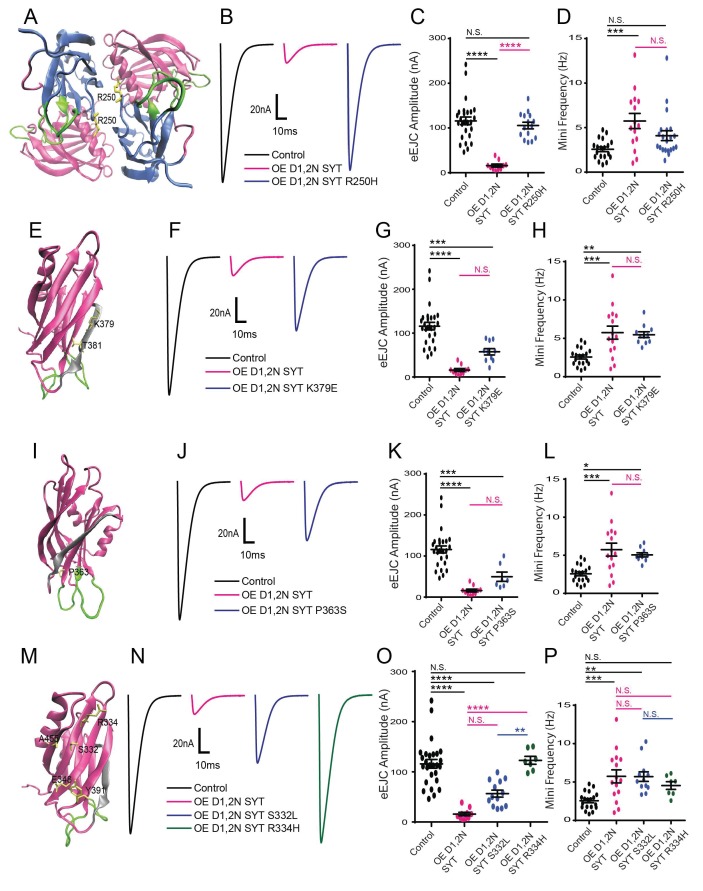
Figure 2. Characterization of suppressor mutation effects on DN Syt1C2BD1,2N physiology.
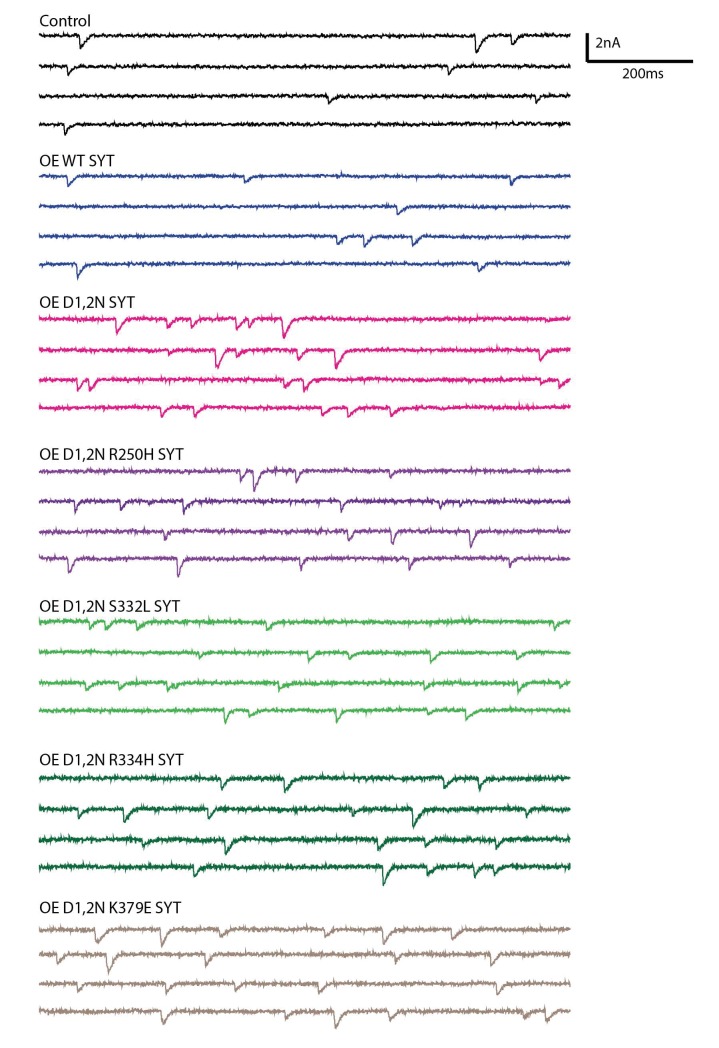
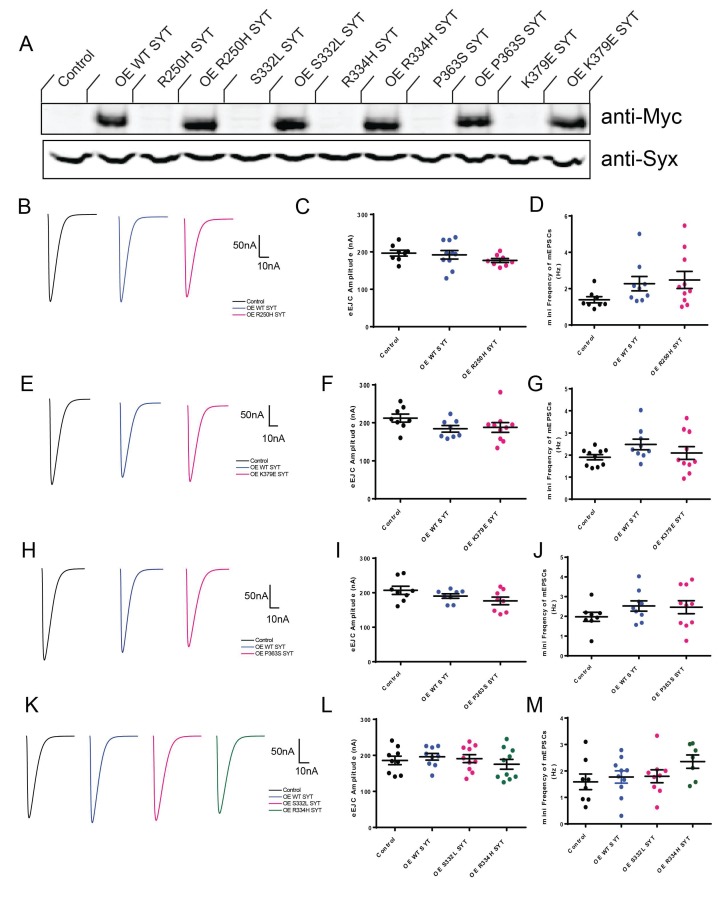
(A) Location of the R250 residue (yellow) in C2A at the Syt1 dimer interface. (B) Representative eEJCs recorded in 0.2 mM extracellular Ca2+ in control (black), elavC155-GAL4; UAS-Syt1C2BD1,2N (OE D1,2N SYT, magenta) and elavC155-GAL4; UAS-Syt1C2BD1,2N R250H (OE D1,2 N SYT R250H, blue). (C) Quantification of mean eEJC amplitude for the indicated genotypes: control, 116.0 ± 8.7 nA, n = 27; OE D1,2N Syt1, 15.9 ± 3.5 nA, n = 10; OE D1,2 N Syt1 R250H, 105.5 ± 7.5 nA, n = 15. (D) Quantification of average mini frequency for the indicated genotypes: control, 2.6 ± 0.3 Hz, n = 19; OE D1,2N Syt1, 5.7 ± 0.8 Hz, n = 15; OE D1,2N Syt1 R250H, 4.1 ± 0.6 Hz, n = 21. (E) Location of the K379 and T381 residues (yellow) in the C2B polybasic β-strand (grey). (F) Representative eEJCs recorded in 0.2 mM extracellular Ca2+ in control (black), elavC155-GAL4; UAS-Syt1C2BD1,2N (OE D1,2N SYT in magenta) and elavC155-GAL4; UAS-Syt1C2BD1,2N K379E (OE D1,2N SYT K379E, blue). (G) Quantification of mean eEJC amplitude in the indicated genotypes: control, 116.0 ± 8.7 nA, n = 27; OE D1,2N Syt1, 15.9 ± 3.5 nA, n = 10; OE D1,2N Syt1 K379E, 57.7 ± 7.3 nA, n = 10. (H) Quantification of average mini frequency for the indicated genotypes: control, 2.6 ± 0.3 Hz, n = 19; OE D1,2N Syt1, 5.7 ± 0.8 Hz, n = 15; OE D1,2N Syt1 K379E, 5.5 ± 0.4 Hz, n = 11. (I) Location of the P363 residue (yellow) near the C2B Ca2+ binding loops (green). (J) Representative eEJCs recorded in 0.2 mM extracellular Ca2+ in control (black), elavC155-GAL4; UAS-Syt1C2BD1,2N (OE D1,2N SYT, magenta) and elavC155-GAL4; UAS-Syt1C2BD1,2N K379E (OE D1,2N SYT P363S, blue). (K) Quantification of mean eEJC amplitude in the indicated genotypes: control, 116.0 ± 8.7 nA, n = 27; OE D1,2N Syt1, 15.9 ± 3.5 nA, n = 10; OE D1,2N Syt1 P363S, 49.6 ± 11.2 nA, n = 7. (L) Quantification of average mini frequency for the indicated genotypes: control, 2.6 ± 0.3 Hz, n = 19; OE D1,2N Syt1, 5.7 ± 0.8 Hz, n = 15; OE D1,2N Syt1 P363S, 5.1 ± 0.3 Hz, n = 10. (M) Location of the S332, R334, E348, Y391, and A455 residues (yellow) on the C2B surface opposite to the polybasic β-strand (grey). (N) Representative eEJCs recorded in 0.2 mM extracellular Ca2+ in control (black), elavC155-GAL4; UAS-Syt1C2BD1,2N (OE D1,2N SYT, magenta), elavC155-GAL4; UAS-Syt1C2BD1,2N S332L (OE D1,2N SYT S332L, blue) and elavC155-GAL4; UAS-Syt1C2BD1,2N R334H (OE D1,2N SYT R334H, green). (O) Quantification of mean eEJC amplitude in the indicated genotypes: control, 116.0 ± 8.7 nA, n = 27; OE D1,2N Syt1, 15.9 ± 3.5 nA, n = 10; OE D1,2N Syt1 S332L, 56.6 ± 7.0 nA, n = 13; OE D1,2N Syt1 R334H, 122.9 ± 7.8 nA, n = 7. (P) Quantification of average mini frequency for the indicated genotypes: control, 2.6 ± 0.3 Hz, n = 19; OE D1,2N Syt1, 5.7 ± 0.8 Hz, n = 15; OE D1,2N Syt1 S332L, 5.7 ± 0.6 Hz, n = 12; OE D1,2N Syt1 R334H, 4.5 ± 0.5 Hz, n = 7. Statistical significance was determined using one-way ANOVA (nonparametric) with post hoc Sidak’s multiple comparisons test. N.S. = no significant change, *p<0.05, **=p<0.005, ***=p<0.0005, ****=p<0.0001. All error bars are SEM.