Abstract
P-element–induced wimpy testes (Piwi) proteins are known for suppressing retrotransposon activation in the mammalian germline. However, whether Piwi protein or Piwi-dependent functions occur in the mammalian soma is unclear. Contrary to germline-restricted expression, we observed that Piwi-like Miwi2 mRNA is indeed expressed in epithelial cells of the lung in adult mice and that it is induced during pneumonia. Further investigation revealed that MIWI2 protein localized to the cytoplasm of a discrete population of multiciliated airway epithelial cells. Isolation and next-generation sequencing of MIWI2-positive multiciliated cells revealed that they are phenotypically distinct from neighboring MIWI2-negative multiciliated cells. Mice lacking MIWI2 exhibited an altered balance of airway epithelial cells, demonstrating fewer multiciliated cells and an increase in club cells. During pneumococcal pneumonia, Miwi2-deficient mice exhibited increased expression of inflammatory mediators and increased immune cell recruitment, leading to enhanced bacterial clearance. Taken together, our data delineate MIWI2-dependent functions outside of the germline and demonstrate the presence of distinct subsets of airway multiciliated cells that can be discriminated by MIWI2 expression. By demonstrating roles for MIWI2 in airway cell identity and pulmonary innate immunity, these studies elucidate unanticipated physiological functions for Piwi proteins in somatic tissues.
Keywords: Cell Biology, Pulmonology
Keywords: Innate immunity
Introduction
The lung epithelium is composed of a variety of cellular lineages that reside in anatomically specific niches from the trachea to the conducting airways to the alveoli. These epithelial lineages are highly specialized to function in both physiological and immunological capacities to maintain pulmonary homeostasis (1). Delineation of the diverse cellular subtypes residing in the pulmonary epithelium is rapidly progressing due to technological advances including genome-wide transcriptional profiling and single-cell sequencing. These studies have revealed a more complex population of lung epithelial cells than had been previously appreciated (2). A recent study from our group revealed the unanticipated finding that epithelial cells from lungs with bacterial pneumonia exhibited higher expression of the mRNA encoding the Piwi protein MIWI2 during pneumococcal pneumonia (3).
Piwi family proteins have been identified in evolutionarily distant species from sponges to humans (4), and their name, P-element–induced wimpy testes, is reflective of the initial investigations elucidating their functional role in the male germline. In germ cells, MIWI2 binds a distinct class of small RNAs designated as Piwi-interacting RNAs (piRNAs), through which they silence the expression of retroelements in the genome (5, 6). Mice deficient in one or all of the Piwi proteins are viable, yet display defects in spermatogenesis (5, 7–9). Outside of the germline, Piwi protein expression has been detected in certain human cancer cells, and may be linked to invasion and metastasis, or serve as a potential prognostic marker (10, 11). Additionally, the expression of Piwi proteins has also been reported in hematopoietic stem cells (12, 13), but focused investigations of potential somatic roles for MIWI2 in the context of hematopoietic development (9, 14) and pluripotency (15) have been unable to identify any extragonadal function in mammals.
In the present study, we investigated MIWI2 expression in the lungs. We found that a subpopulation of multiciliated airway epithelial cells express MIWI2. Both the number of MIWI2-positive cells and the degree to which they express this factor increase during lung infection. RNA sequencing demonstrated that MIWI2-positive cells have a significantly different transcriptome compared with neighboring multiciliated cells that do not express MIWI2. Furthermore, MIWI2 loss-of-function studies revealed a functional role for MIWI2 in the homeostatic balance of ciliated and secretory cells in the conducting airways and in shaping innate immune responses. Overall, these results revealed that multiciliated cells, which have previously been thought to be homogenous, can be reclassified into MIWI2-positive and MIWI2-negative subsets. More broadly, they provide the first evidence that Piwi proteins are functional in noncancerous somatic tissue.
Results
MIWI2 is expressed in adult lungs and induced during pneumococcal infection.
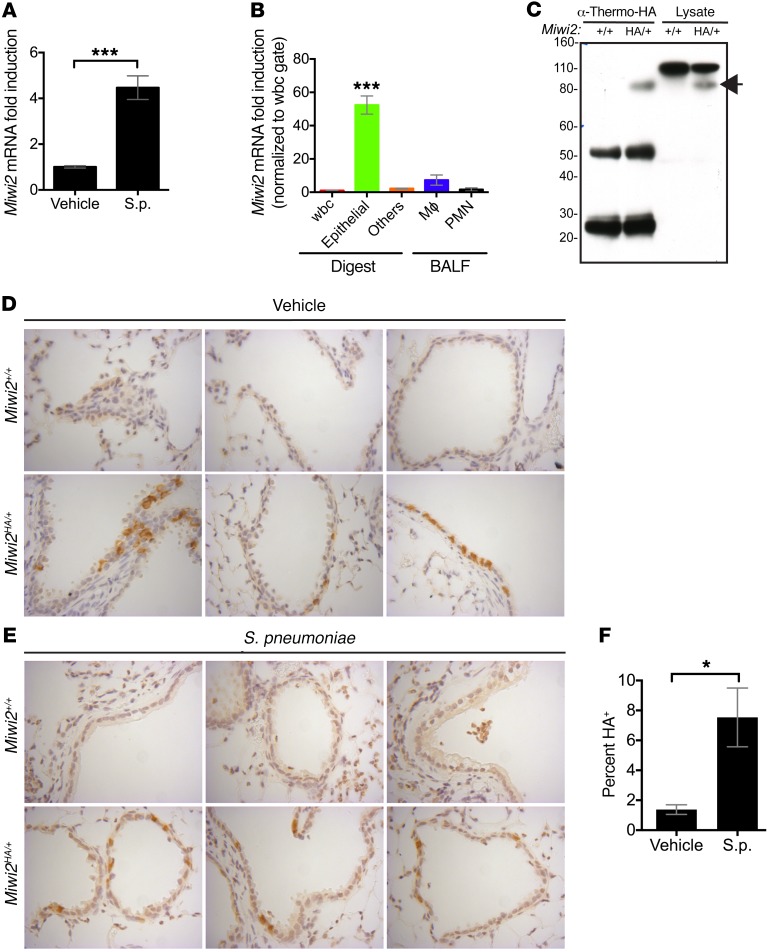
In a previous genomic profiling study, we investigated genes that were increased in lung cell populations in a clinically relevant murine model of Streptococcus pneumoniae pneumonia (3). Among the mRNAs significantly induced during lung infection was the transcript encoding Miwi2. This was particularly notable given that this protein is thought to function exclusively in vertebrate male germ cells, not in lungs nor any other somatic tissues. To corroborate the profiling study findings, we confirmed that Miwi2 mRNA expression was increased approximately 3-fold in lung epithelial cells during infection using quantitative real-time PCR (Figure 1A).
Figure 1. Lung epithelial cells express MIWI2.
(A) Quantitative real-time PCR of epithelial RNA isolated from mice infected intratracheally (i.t.) with S. pneumoniae. Data represent fold induction over vehicle-treated mice, presented as mean ± SEM, n = 7 mice per group. ***P < 0.001 as determined by unpaired t test. (B) Quantitative real-time PCR of RNA isolated from FACS-sorted cells from mice infected i.t. with S. pneumoniae. Data represent fold induction over leukocyte expression, presented as mean ± SEM, n = 6 mice per group. ***P < 0.001 as determined by 1-way ANOVA, followed by Tukey post hoc test. (C) Immunoblot analysis for HA-MIWI2 after immunoprecipitation with anti-HA antibody, or whole lung lysate from Miwi2+/+ or Miwi2HA/+ mice. Arrow indicates band at expected size of MIWI2 (98 kDa). Image is representative of an experiment performed 3 times. (D and E) Immunostaining with anti-HA antibody of lung sections from Miwi2+/+ or Miwi2HA/+ mice treated with vehicle (D) or infected i.t. with S. pneumoniae (E). Images were acquired using the ×40 objective. (F) Morphometric quantitation was performed by counting of the number of MIWI2-positive cells per airway. At least 3 fields were analyzed from 2 sections per mouse, n = 4 mice per group, presented as mean ± SEM. *P < 0.05 as determined by unpaired t test.
Specific cell sources of Miwi2 expression were interrogated in mice infected with S. pneumoniae using a previously described protocol for isolating purified cell populations recovered from whole lung enzymatic digests and bronchoalveolar lavage fluid (3, 16). Relative abundance of Miwi2 transcript was assessed by quantitative real-time PCR and determined to be enriched in the EpCAM+CD45– population, indicating that cells within the epithelium are particularly enriched for lung Miwi2 mRNA expression (Figure 1B).
Given that steady-state mRNA levels are not necessarily accurate surrogates of corresponding protein abundance (17), MIWI2 protein analyses were conducted. We found that readily available antibodies against mouse MIWI2 were unreliable in tissue sections. Thus, we used a novel MIWI2-fusion mouse model engineered with an in-frame influenza virus hemagglutinin (HA) tag inserted immediately downstream of the endogenous Miwi2 start codon to assess MIWI2 protein expression (18). No fertility defects were observed in Miwi2HA/HA mice, indicating that the HA-MIWI2 fusion protein is expressed normally and does not disrupt endogenous MIWI2 function. Immunoprecipitation and Western blot of whole lung lysates from Miwi2HA/+ or Miwi2+/+ mice demonstrated that full-length MIWI2 protein is expressed in the lungs (Figure 1C).
Our previous studies indicated that EpCAM+CD45– cells from whole lung digests contain the major pulmonary epithelial subtypes, including club cells, multiciliated cells, and type I and II pneumocytes (16). We evaluated lung sections by immunohistochemistry to determine the cell type and anatomic location of MIWI2 protein expression in the lungs. Immunohistochemical analysis of lung sections from uninfected Miwi2HA/+ mice revealed that MIWI2 expression was cytoplasmic and restricted to the proximal and conducting airway epithelium, with no detectable expression in the distal alveolar epithelium (Figure 1D). After infection with S. pneumoniae, MIWI2 expression was still localized in the cytoplasm, and restricted to airway cells (Figure 1E). To determine whether the increase in Miwi2 mRNA correlated with increased MIWI2 protein expression, tissue sections isolated from uninfected or infected lungs were quantitatively compared, and demonstrated that the number of MIWI2-positive cells was significantly increased as a result of infection (Figure 1F).
MIWI2 marks a subpopulation of multiciliated cells in both mouse and human airway.
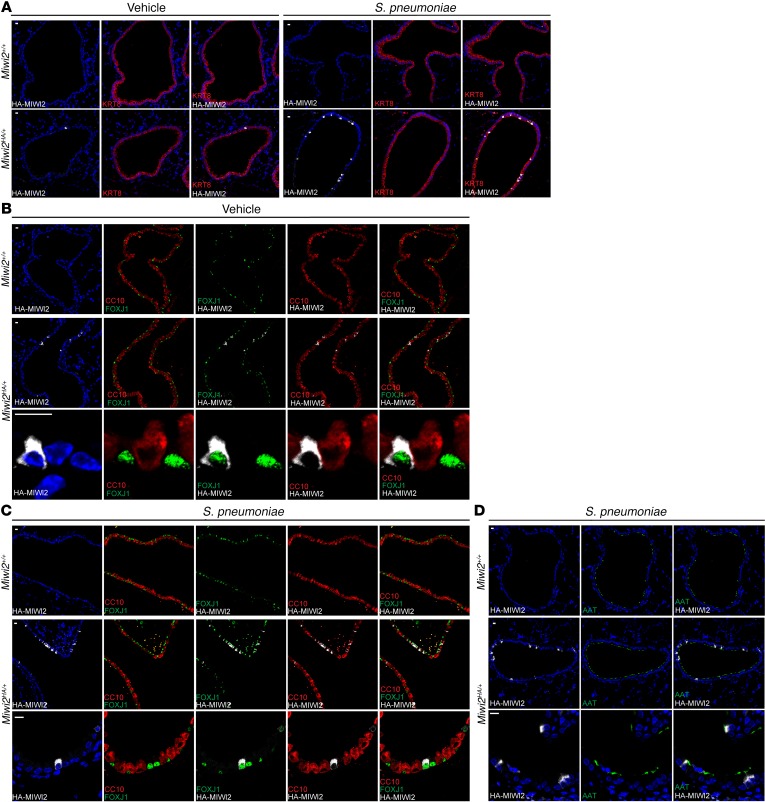
As the airway epithelium in mice and humans is composed of several cell types, we sought to determine which epithelial cell population(s) expressed MIWI2. Furthermore, we sought to determine whether similar or differing cell types express MIWI2 after infection. Immunofluorescence staining indicated that MIWI2 expression was exclusively restricted to airway luminal cells expressing keratin 8 (KRT8) (Figure 2A), and excluded from KRT5-expressing basal cells (data not shown). In both infected and uninfected lungs, all MIWI2-positive cells were marked by FOXJ1, but not all FOXJ1+ cells expressed MIWI2, suggesting that MIWI2 distinguished a subset of multiciliated cells (Figure 2, B and C). These conclusions were validated using a separate and independent marker of ciliated cells, acetylated α-tubulin (Figure 2D), which confirms that MIWI2-positive cells express multicilia.
Figure 2. MIWI2 marks a subpopulation of multiciliated cells.
(A) Immunostaining for KRT8 (red) and HA epitope (white) of lung sections from Miwi2+/+ or Miwi2HA/+ mice treated with vehicle or infected i.t. with S. pneumoniae. (B and C) Immunostaining for CC10 (red), HA epitope (white), FOXJ1 (green), or Hoechst (blue) on paraffin-embedded lung sections from vehicle-treated (B) or S. pneumoniae–infected (C) Miwi2+/+ or Miwi2HA/+ mice. (D) Immunostaining for acetylated α-tubulin (green) and HA epitope (white) of lung sections from Miwi2+/+ or Miwi2HA/+ mice infected i.t. with S. pneumoniae. Representative results are shown from staining performed on at least 2 sections, n = 6 mice per group. Scale bars: 10 μm.
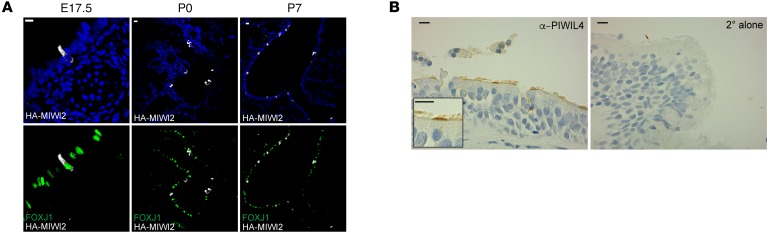
To determine homeostatic MIWI2 expression outside of exposure to pathogens or inhaled particles, we evaluated its content in lung sections from mice collected before the first breath at E17.5. Additionally, we examined MIWI2 expression at P0 and P7. In agreement with data collected from adult lungs, MIWI2 expression was localized exclusively to conducting airways and excluded from distal airways. As early as E17.5, 100% of MIWI2-positive cells also expressed FOXJ1 (Figure 3A).
Figure 3. MIWI2 is expressed in ciliated cells of the embryonic and human lung.
(A) Immunostaining for HA epitope (white) and FOXJ1 (green) on paraffin-embedded lung sections from Miwi2HA/+ mice collected from timed pregnancies. Representative results are shown from staining performed on at least 2 sections, n = 4 mice per group. Scale bars: 10 μm. (B) Immunohistochemical analysis of human lung sections stained with anti-PIWIL4 antibody or secondary antibody alone. Image is representative of similar findings from 3 separate human donors. Scale bars: 10 μm.
Because of the differences between mouse and human cellular airway composition, we investigated whether MIWI2-positive ciliated cells exist in human airways. Lung sections from 3 separate normal donors were stained with an antibody against the human homolog of MIWI2, PIWIL4 (also known as HIWI2). In agreement with our findings derived from murine lungs, a discrete subset of multiciliated cells expressed PIWIL4 (Figure 3B), although, unlike our findings in mice, staining in human cells was enriched in the apical membrane region where basal bodies reside. This could represent species differences in intracellular MIWI2 localization, or instead reflect an effect of the epitope HA tag used in the murine model. Collectively, these data show that MIWI2 protein was selectively expressed outside of the germline in a subset of lung ciliated epithelial cells in both mice and humans. Furthermore, during respiratory infection, the number of MIWI2-positive multiciliated cells increased and MIWI2 remained cytoplasmically localized in airway epithelial cells.
MIWI2-positive epithelial cells represent a distinct subset of multiciliated cells.
Multiciliated cells in the airways are terminally differentiated. They arise from at least 2 different sources: KRT5+ basal cells and club cells (19, 20). Despite varied origins, current evidence supports the concept that the differentiated multiciliated cell population is homogenous (19, 21, 22). Other major lung cell lineages, including club cells, type I cells, and type II cells, exhibit functionally distinct subtypes (23, 24), but to our knowledge no phenotypic variation has been defined for airway multiciliated cells. MIWI2 expression in a subset of multiciliated cells led us to question the commonly held notion that multiciliated cells are homogenous.
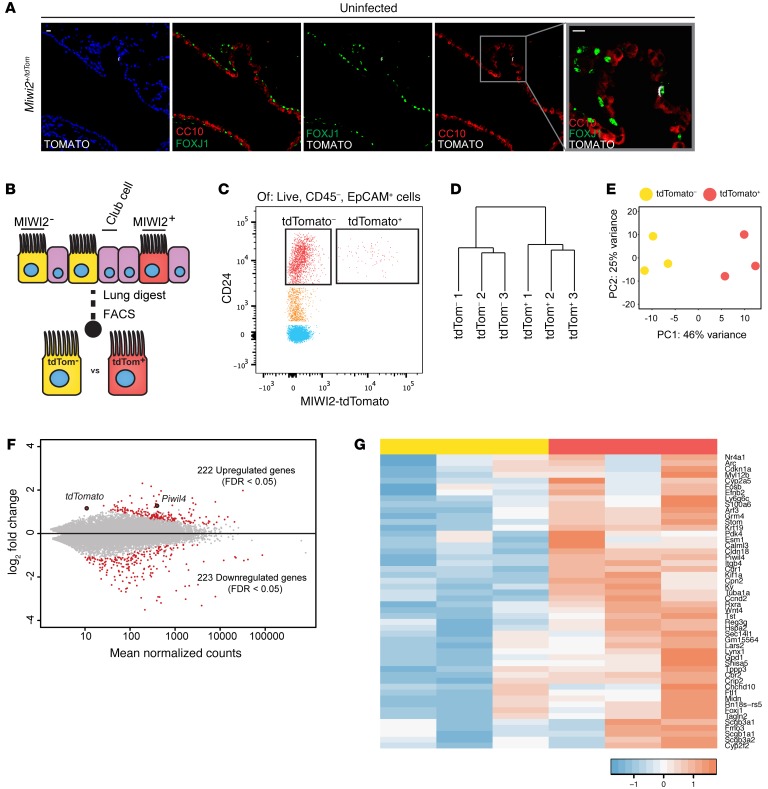
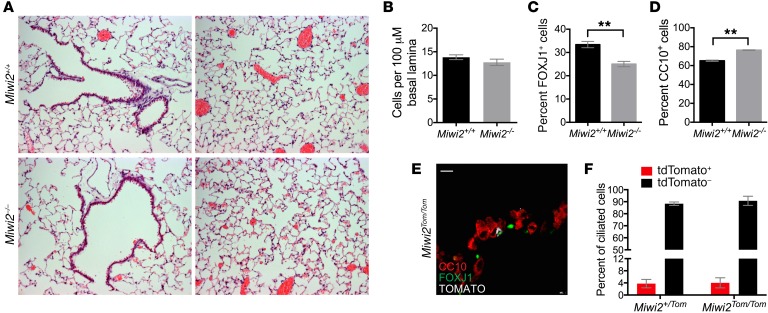
As MIWI2 was exclusively localized to a subset of multiciliated cells, we determined whether MIWI2-positive multiciliated cells express a transcriptional program that is distinct from that of MIWI2-negative multiciliated cells. Using a MIWI2 reporter model (Miwi2+/Tom) whereby a tdTomato expression cassette was recombined into the endogenous Miwi2 allele at the first coding exon (25), we observed that tdTomato expression was restricted to a subset of ciliated cells (Figure 4A). This both confirmed the cellular localization of MIWI2 expression in the lung and validated this animal model for tracking MIWI2 transcript expression. Multiciliated cells were isolated by FACS from enzymatic lung digests (Figure 4B) based on EpCAM and CD24 expression, a strategy previously described by others (26) and independently validated in our current studies (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI94639DS1). Using the organ-wide and more exhaustive analyses of lung cells afforded by FACS, all MIWI2-expressing cells (based on the tdTomato reporter) were within the CD45–EpCAM+CD24hi cell population (Figure 4C), providing yet another complementary line of evidence demonstrating the pulmonary expression of this gene exclusively within a subset of airway multiciliated cells.
Figure 4. MIWI2-positive airway cells represent a transcriptionally discrete subpopulation of multiciliated cells.
(A) Representative lung sections from Miwi2+/Tom mice immunostained for red fluorescent protein (RFP, white), FOXJ1 (green), and CC10 (red). Scale bars: 10 μm. (B) MIWI2+ cells are labeled through the expression of tdTomato. (C) Representative cytometry plot selected for live, CD45–, EpCAM+ cells from uninfected Miwi2+/Tom mouse after elastase lung digest. (D) Unsupervised hierarchical clustering based on whole transcriptome data from MIWI2+ and MIWI2– ciliated cells. (E) Principal component analysis of the data sets analyzed in C. (F) MA plot representation of DESeq output. Red dots indicate genes determined to be significant at FDR q < 0.05. Piwil4 (an alias of Miwi2) and tdTomato are indicated. (G) Heatmap representation of top 50 genes induced in MIWI2-positive versus MIWI2-negative ciliated cells. Values normalized by Z score. Data presented are from n = 3 mice per group.
To identify whether transcriptional profiles differ between these subsets of multiciliated cells, we used the tdTomato signal to sort CD45–EpCAM+CD24hi cells from uninfected lungs into MIWI2-positive and MIWI2-negative populations, upon which we performed RNA-Seq analyses. The transcriptomes significantly differed between these 2 cellular cohorts, as depicted by unsupervised hierarchical clustering and principal component analysis (Figure 4, D and E). Transcripts from more than 400 genes significantly differed between groups with stringency of FDR <0.05, with about half enriched in the MIWI2-positive population while the other half were downregulated (Figure 4F). The 50 most significantly enriched genes in MIWI2-positive cells are illustrated in Figure 4G. Further analysis of the 222 genes enriched in MIWI2-positive cells revealed that 20 (9%) have been previously identified as specific markers of club cells, including the hallmark transcripts for secretoglobins Scgb1a1(encoding CC10) and Scgb3a2 (2). Club cells are nonciliated and produce many secretory products that are protective for the maintenance of normal lung homeostasis. Together with basal cells, club cells are multipotent progenitors capable of differentiating into airway multiciliated cells (22, 27). Despite the presence of club cell–like mRNAs, we did not observe a single instance of a cell coexpressing MIWI2 and CC10 protein in the immunofluorescence analyses.
MIWI2 influences airway epithelial layer composition.
The restricted expression of MIWI2 in a subset of ciliated cells and its induction during pneumonia led us to determine whether MIWI2 expression impacts airway epithelial cell composition. To investigate whether MIWI2 could play a role in mediating the composition of airway epithelial cells, we examined histological sections of Miwi2+/+ and Miwi2–/– mice (Figure 5A). No gross differences were appreciated between Miwi2+/+ and Miwi2–/– mice. Further investigation of airway cell composition was carried out using immunofluorescence. Although the total airway cell numbers were equivalent between genotypes, as determined by morphometric quantification of all nucleated cells (Figure 5B), loss of MIWI2 reduced the number of ciliated cells and led to a concomitant increase in total club cells (Figure 5, C and D). This presents the interesting hypothesis that MIWI2 protein function is required for the generation of the MIWI2-positive cell lineage. To test this, we leveraged the Miwi2-tdTomato reporter mice, in which the Miwi2 locus is replaced by a tdTomato-expressing null allele (25). If MIWI2 protein was required for the generation of MIWI2-positive cells, we would have expected to observe no ciliated cells expressing the tdTomato reporter. Instead, examination of both tissue sections and flow cytometry of lung digests generated from Miwi2Tom/Tom mice demonstrated the presence of Miwi2-tdTomato+ cells (Figure 5E). Furthermore, the percentage of ciliated cells with an active Miwi2 promoter (Miwi2Tom/Tom) was unchanged in comparison with a mouse with a single functional allele of Miwi2 (Miwi2+/Tom) (Figure 5F). Collectively these data lead to the conclusion that MIWI2 alters the balance between multiciliated and club cell populations in the airways, but MIWI2 protein itself is not required for the generation of the distinct subsets of MIWI2-positive and -negative multiciliated cells.
Figure 5. MIWI2 regulates pulmonary epithelial cell composition.
(A) H&E staining of lung sections from uninfected Miwi2+/+ and Miwi2–/– mice. (B–D) Lung sections from uninfected Miwi2+/+ and Miwi2–/– mice were immunostained for CC10, FOXJ1, and DAPI, and the total numbers of airway cells (B), ciliated cells (C), and club cells (D) were quantified. n = 4, 3 mice per group, data presented as mean ± SEM. **P < 0.01 as determined by unpaired t test. (E) Lung sections from Miwi2Tom/Tom mice immunostained for red fluorescent protein (RFP, white), FOXJ1 (green), and CC10 (red). Scale bars: 10 μm. (F) Flow cytometric quantification of tdTomato-positive and tdTomato-negative cells as a percentage of live, CD45–EpCAM+CD24hi ciliated cells; n = 3, 4 mice per group, data presented as mean ± SEM.
MIWI2 regulates lung innate immune responses.
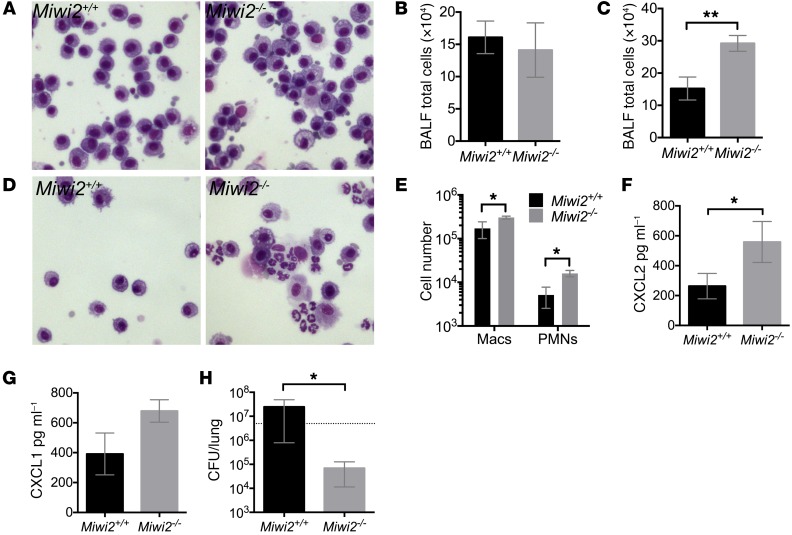
In addition to playing a homeostatic role in determining the cell composition of the pulmonary epithelial barrier, MIWI2 is induced during bacterial pneumonia. Therefore, we investigated whether MIWI2 impacts lung innate immunity. We infected Miwi2+/+ and Miwi2–/– mice with S. pneumoniae, the most common bacterial cause of community-acquired pneumonia. No differences were detected in airspace cellularity at baseline in the absence of infection (Figure 6, A and B). Four hours after an intratracheal instillation of bacteria, a time point at which leukocyte emigration to the airspace is first appreciable, a greater number of cells was recovered in bronchoalveolar lavage fluid (BALF) from Miwi2–/– mice (Figure 6C), owing to significant increases in both macrophages and neutrophils (Figure 6, D and E). Enhanced leukocyte recruitment was accompanied by increased levels of the neutrophil chemokine CXCL2 (Figure 6F), with a similar, albeit statistically insignificant, trend for CXCL1 (Figure 6G). To determine whether these markers of an enhanced innate immune response were robust enough to impact host defense, we harvested lung tissue 30 hours after bacterial instillation, to allow time for immunity to impact infection. Miwi2–/– mice showed a significant reduction in pulmonary bacterial burden (Figure 6H). Taken together, these data demonstrate that MIWI2 regulates pulmonary inflammation and defense, likely because of its presence in the pulmonary epithelial cell barrier of the respiratory tract.
Figure 6. MIWI2 regulates pulmonary innate immunity during pneumonia.
(A and B) BALF cell cytospins (A) and total BALF cell counts (B) of uninfected Miwi2+/+ and Miwi2–/– mice. n = 4, 4 mice per group, data are expressed as mean ± SEM. Microscopic images were obtained using the ×40 objective. (C–G) Total BALF cell counts (C), BALF cytospins (D), BALF differential counts (E), BALF CXCL2 protein (F), and BALF CXCL1 protein (G) were measured from Miwi2+/+ and Miwi2–/– mice infected i.t. with S. pneumoniae for 4 hours. n = 6, 7 mice per group, data are expressed as mean ± SEM. *P < 0.05, **P < 0.01 as determined by unpaired t test. Microscopic images were obtained using the ×40 objective. (H) Lung CFU was determined in Miwi2+/+ and Miwi2–/– mice infected i.t. with S. pneumoniae and harvested 30 hours later. n = 6, 9 mice per group, data represented as mean ± SEM. *P < 0.05 as determined by unpaired t test. Dotted line indicates input CFU.
Discussion
In germ cells, Piwi proteins function during meiosis to defend the genome from aberrant retrotransposon expression and potential reintegration. Despite the primary focus on Piwi-clade proteins in the germline, prior evidence also suggested that these proteins could also be expressed in somatic cells. Not long after the first identification of Piwi as a critical factor for stem cell renewal in germline stem cells, PIWI was also observed in the supporting somatic cells of the Drosophila ovary (28) and in the testicular somatic cells (25, 29). Reports of Piwi-clade mRNA expression in mammals have remained focused on stem and cancer cell populations. While mRNA expression has been detected, neither protein-level expression nor functional significance has ever yet been identified for Piwi proteins outside of the germline in mammals. Transcripts for HIWI, the human homolog of Drosophila Piwi, and for mouse Miwi are detected in CD34+ hematopoietic progenitor cells and are downregulated upon differentiation to mature myeloid cells (12). These data suggested that Piwi proteins might play a role in maintaining stem cell properties in somatic cells in mammals, similar to their role in the Drosophila ovary. However, mice deficient in all 3 Piwi-clade proteins, Miwi, Mili, and Miwi2, exhibit neither dysregulation of hematopoiesis nor aberrations in progenitor cell gene expression (9, 14). In addition, induced pluripotent stem cells generated from Miwi/Mili/Miwi2-triple-knockout mice differentiate into all 3 germ layers, indicating that these PIWI proteins are collectively dispensable for stem cell differentiation and maintenance in mice (15). In the present study, we report that MIWI2 is expressed at the protein level in somatic cells, specifically in a distinct subset of lung epithelial cells.
Several types of lung epithelial cells are heterogeneous in their gene expression profiles. In contrast to club cells and type II cells, heterogeneity within airway multiciliated cells has not been identified. Our data indicate that a small proportion of multiciliated cells express MIWI2 as early as E17.5, and that the number of these MIWI2-positive cells is increased during infection with pneumococcus. MIWI2-positive ciliated cells were apparent throughout the bronchial tree, from trachea to conducting airways; however, no association was noted with any specific anatomical feature, nor were MIWI2-positive cells juxtaposed with pulmonary neuroendocrine cells, which constitute neuroepithelial bodies (ref. 30 and data not shown). While all MIWI2-positive cells expressed the ciliated marker FOXJ1, their transcriptional profiles were substantially altered compared with those of MIWI2-negative ciliated cells. The results demonstrate that there are at least 2 different subpopulations of multiciliated cells in the pulmonary epithelium. Ciliated cells expressing MIWI2 have a phenotype characterized by a distinct transcriptome from their neighboring ciliated cells that do not express this Piwi protein.
The RNA-Seq analyses also revealed that MIWI2-postive ciliated cells have an increase in club cell transcripts, suggesting the intriguing new hypothesis that MIWI2-positive cells could be multiciliated cells recently derived from club cells. It is conceivable that the MIWI2-positive ciliated cells have secreted their protein cargo (since club cell markers are secretory proteins and not observed in any of the MIWI2-positive cells) yet retain some of the transcripts from their forebear cells. The transcriptional signature may thus reveal a recent cellular history. Additional lineage-tracing studies will be needed to conclusively identify cellular origins and test the hypothesis here suggested that MIWI2-positive airway multiciliated cells may selectively derive from club cells.
Our data demonstrate that MIWI2 in airway cells is observed in gestational, neonatal, and adult ages, restricted to the cytoplasm in all cases. These expression characteristics differ markedly from MIWI2 expression in the germline. MIWI2 protein expression in the male germline occurs during prepachytene, beginning at E15.5 and absent by P3 (31, 32). In these germ cells, the MIWI2 protein is primarily localized to the nucleus, although some perinuclear staining is also evident (31, 32). The nuclear localization depends on piRNA loading of MIWI2 by another Piwi protein (18, 31). Our data demonstrate that, in the lungs, MIWI2 is expressed prenatally through adulthood and is predominantly or even exclusively found in the cytoplasm. These disparate patterns of expression suggest that the regulation and function of MIWI2 in somatic cells may be radically different from in the germline.
For the first time to our knowledge, our results using a model of MIWI2-deficiency reveal a somatic cell function for Piwi family proteins. During homeostasis, Miwi2–/– mice exhibited altered airway cell composition, noted by a decrease in ciliated cells and an increase in club cells. Given that the decrease in ciliated cells in Miwi2–/– mice is similar to the percentage of ciliated cells that are MIWI2-positive, we considered the hypothesis that the epithelial skewing could simply result from the loss of MIWI2-positive ciliated cells. However, Miwi2-tdTomato homozygous mice, which have no functional MIWI2, demonstrate a proportion of tdTomato-expressing cells similar to that shown by Miwi2-tdTomato heterozygous mice, which express MIWI2. Thus, MIWI2 does not appear necessary for establishing heterogeneous subpopulations within the airway ciliated cells. The mechanism by which MIWI2 influences the balance of secretory and ciliated cells remains to be determined. For example, excessive Notch signaling drives secretory cell metaplasia and a loss of ciliated cells (27), so MIWI2 could influence epithelial cellularity if it regulates the expression of Notch or perhaps soluble cell fate determinants.
We initially observed Miwi2 mRNA induction while examining the epithelial response to acute bacterial pneumonia (3), and our data now indicate that MIWI2 regulates innate immunity during pulmonary infection. MIWI2 deficiency results in an increased early recruitment of neutrophils to the airspace, and increased bacterial clearance. It is conceivable that some of these tissue-protective effects may be attributable to differences in airway epithelial cell composition. Transgenic mice that are engineered to overexpress the mucin gene Muc5b in club cells exhibited an increase in inflammation and immune cell recruitment when infected with a Gram-positive pathogen (33). This was not merely a direct antibacterial effect, but rather attributable, in part, to improved alveolar macrophage function. Interestingly, we observed an increase in CXCL2 expression in Miwi2–/– mice, which is derived exclusively from macrophages during early stages of pneumococcal pneumonia (34). While secretory metaplasia and hypersecretory phenotypes are pathological in many lung diseases (35), such as cystic fibrosis and asthma, it is conceivable that small increases in club cell numbers may impart improved antibacterial defenses during infection. For instance, several studies have shown that club cells contribute to acute pulmonary inflammation in response to bacterial stimuli (36, 37), which is consistent with our own observations of increased club cells in MIWI2-deficient mice with pneumonia. Additionally, the localization of PIWIL4 (also known as HIWI2) at the apical surface of the ciliated cell suggests the possibility MIWI2 may interact with cilia machinery, which could in turn influence mucociliary clearance. While Miwi2Tom/Tom cells, which are functionally deficient in MIWI2, express multicilia, the influence on ciliary function has not been explored.
A limitation to the functional analyses in the present studies was their reliance on a total-body knockout of Miwi2. As discussed previously, MIWI2 was thought to be expressed only in the germline perinatally in males. We have identified somatic expression in lung epithelium; however, the possibility exists that MIWI2 is expressed in other cell types in the lung, at levels undetectable by HA immunostaining or tdTomato expression. Importantly, while the germline phenotype of Miwi2–/– mice is limited to male mice (5), our studies used both male and female mice. Nonetheless, future studies interrogating the mechanisms of MIWI2 function in airway ciliated cells can use cell-specific Cre drivers.
Multiple independent and complementary approaches demonstrate that MIWI2 protein is constitutively and inducibly expressed in noncancerous somatic tissues of mammals, in a subset of multiciliated cells in the lungs of both mice and humans. Furthermore, MIWI2 dictates the balance of secretory and ciliated cells in the lung, and regulates innate immunity during pulmonary infection. While these studies have identified unanticipated sites and functions of MIWI2, they raise many important new questions about MIWI2 biology. In addition, by presenting the first evidence that lung ciliated cells represent a heterogeneous rather than homogenous population of cells, these results guide new lines of investigation into structure-function relationships in the respiratory tract. These studies provide new insights and directions for pulmonology, immunology, and Piwi protein biology by demonstrating cell specification and innate immunity roles of MIWI2 in multiciliated cells of the lung.
Methods
Mice and human samples.
Mice were maintained under pathogen-free conditions, under a 12-hour light-dark cycle, with access to food and water ad libitum. All experiments were performed using male and female mice at 8–20 weeks of age, unless otherwise indicated. C57BL/6 mice were obtained from The Jackson Laboratory. The Miwi2–/– mice were provided by Gregory Hannon, and have been described previously (5). These mice were rederived at the Boston University School of Medicine transgenic core, and bred as Miwi2+/– × Miwi2+/– crosses to generate littermate controls. The Miwi2HA/+ and Miwi2tdTomato/+ mice were described previously (18, 25). The Miwi2HA/+ mice were backcrossed for more than 6 generations to C57BL/6 and maintained as Miwi2HA/+ × C57BL/6. All mouse experiments were performed at least twice, on separate days using mice of different litters. At least 1 control and 1 experimental mouse were included on each day. The total number of mice used for each experiment is presented in the figure legends. For immunohistological studies in human samples, lungs of deidentified adult donors were procured through the International Institute for the Advancement of Medicine (Edison, New Jersey, USA).
Experimental infections.
Experimental pneumonias were generated via the intratracheal route as previously described (38). Mice were anesthetized by an i.p. injection of ketamine (50 mg/kg) and xylazine (5 mg/kg) diluted in sterile saline. The tracheas were cannulated using a 24-gauge angiocatheter that was directed to the left bronchus. A 50-μl bolus of pneumococcus in sterile saline was instilled. Mice were placed in the right lateral decubitus position until they were sternally recumbent. A target instillation of 5 × 106 CFU of bacteria was estimated by optical density and verified by quantification of serial dilutions grown on trypticase soy agar supplemented with 5% sheep blood agar plates at 37°C overnight.
Quantitative real-time PCR.
Quantitative real-time PCR was performed on 100 ng of TRIzol-extracted RNA using a CFX96 Real-Time System (Bio-Rad) and TaqMan RNA-to-CT 1-step kit (Life Technologies). Primers and probes for Miwi2 (Mm01144775) and 18S (Mm03928990) rRNA were obtained from Life Technologies. Fold induction was calculated using the 2ΔΔCt method.
Immunoprecipitation and Western blot.
Lungs from uninfected Miwi2+/+ or Miwi2HA/+ mice were harvested and homogenized in lysis buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40) supplemented with a protease inhibitor cocktail (Sigma-Aldrich) and RNaseOUT (Life Technologies) using a rotor stator homogenizer. Ten milligrams of supernatant was preincubated with 100 μl of protein G–coupled magnetic beads (Invitrogen), and immunoprecipitation was carried out with 100 μl of magnetic beads coupled to 12 μg HA antibody (Thermo Fisher Scientific, 26183). Beads were washed 3 times with wash buffer (10 mM Tris, pH 8, 150 mM NaCl, 1 mM MgCl2, 0.1% NP-40) and 40 μg of protein lysate, and a predefined proportion of immunoprecipitate was suspended in 1× lithium dodecyl sulfate sample loading buffer plus reducing agent (Life Technologies) heated for 10 minutes at 70°C before loading onto a 4%–12% Bis-Tris gel. Protein was then transferred onto an Immobilon-P PVDF membrane (Millipore) using the X-Cell Blot II system. Membranes were probed with rabbit anti-HA epitope (Cell Signaling Technology, C29F4) followed by anti-rabbit IgG-HRP (Cell Signaling Technology) and developed with ECLPlus (GE Healthcare) before exposure to film (GE Healthcare).
Lung digestion.
Lung digests for isolation of epithelial cells were performed using elastase, as previously described (3, 16). Briefly, after euthanasia, lungs were perfused with 10 ml HBSS via the right ventricle through the pulmonary artery. Elastase dissolved in RPMI 1640 medium (4.5U, Roche) was instilled through a tracheal cannula followed by 0.5 ml low-melting agarose solution warmed to 55°C. Lungs were immediately covered with ice for 2 minutes to polymerize the agarose and incubated in 2 ml elastase/RPMI for 45 minutes at 37°C. After this incubation, lung lobes were minced in RPMI 1640 medium containing 50% FBS and 100 U/ml DNase I (Qiagen). Lung single-cell suspensions were subjected to FACS. Dead cells were excluded using 7-aminoactinomycin D (BD Pharmingen). PE-Cy7–conjugated rat anti–mouse CD45 (BD Pharmingen), APC-conjugated rat anti–mouse CD326 (eBioscience), APC-Cy7–conjugated rat anti–mouse Ly6G, and PE–eFluor 610–conjugated rat anti–mouse F4/80 (eBioscience) were used to separate leukocyte and epithelial cells. Cell isolation was performed using FACSAria III (BD Biosciences).
Immunohistochemistry and immunofluorescence.
For immunohistochemistry, mouse lungs were fixed in 4% (vol/vol) paraformaldehyde and were embedded in paraffin. Sections (5 μm) were deparaffinized and treated with 3% (vol/vol) hydrogen peroxide in methanol for 30 minutes. Slides were then blocked with 10% (vol/vol) normal donkey serum and incubated with a 1:250 dilution of anti–HA epitope (Cell Signaling Technology, 3294) antibody overnight. Biotinylated donkey anti-rabbit IgG (Jackson ImmunoResearch) was used as a secondary antibody at 1:200 dilution for 1 hour. Bound antibody was detected using a Vectastain ABC peroxidase kit (Vector Laboratories) and visualized using DAB. Slides were counterstained with hematoxylin and imaged using a Zeiss LSM 700 microscope. For immunofluorescence, lungs were perfused with 10 ml HBSS and fixed, embedded, deparaffinized, and blocked as above. The following primary antibodies were used: FOXJ1 (eBioscience, 2A5), CC10 (Santa Cruz Biotechnology, polyclonal), and HA (Cell Signaling Technology, C29F4). Secondary antibodies, anti–mouse AF488, anti–rabbit AF647, and anti–goat AF555, were all obtained from Life Technologies.
Bronchoalveolar lavage.
After euthanasia by isoflurane overdose, the trachea and lungs were removed en bloc. The trachea was cannulated with a 20-gauge blunted stainless steel catheter and secured with silk suture. Serial 1-ml lavage samples were taken with ice-cold PBS. The lavage fluid was centrifuged at 300 g for 5 minutes at 4°C, and cell pellets were used for differential counts. BALF cell counts were performed as previously described (39), with cytocentrifuged slides stained with the Diff-Quik staining kit (Dade Behring) after counting of suspended cells using a hemocytometer. For BALF cytokine measurement, the supernatant of the first 1-ml lavage sample was saved separately at –80°C for protein analysis by ELISA (R&D Systems).
Bacterial burden measurements.
Mice were sacrificed by overdose of isoflurane. The lung lobes were removed individually and homogenized using stainless steel beads (Next Advance) and 300 μl of sterile water supplemented with 1× protease inhibitor (Roche). Homogenates were serially diluted in PBS and plated on sheep blood agar plates. After an overnight incubation at 37°C, colonies were enumerated and expressed as total CFU per lung.
Sample library preparation and mRNA-Seq.
Total RNA was isolated from sorted cell populations using TRIzol and cleaned using the Qiagen RNeasy Mini Kit. RNA was amplified using the Ovation RNA-Seq System V2. Amplified RNA was sheared to 500 bp and then used to prepare Illumina-compatible libraries using the Illumina TruSeq Nano DNA preparation kit. All libraries were sequenced on a HiSeq 2500 High Output to generate 100-nt single-end read data sets. Sample RNA amplification, library preparation, and RNA sequencing were conducted by the Tufts University Core Facility in Genomics (http://tucf-genomics.tufts.edu/).
Global transcriptomic analyses.
Illumina fastq files were quality-trimmed using FastQC and Cutadapt 1.7.1 (40) and subsequently aligned to the GRCm38.p4 build of the mouse genome (https://www.gencodegenes.org/) using STAR 2.4.2a (41). Only uniquely aligned reads, which accounted for 90%–92% of all reads from the sample set, were used for downstream analysis. Indexed bam files were then converted to raw gene counts using HTSeq (42) using the GENCODE M10 gene annotation reference. The Bioconductor package DESeq2 (43) was used to determine statistical evaluations of expression differences between groups. Read statistics and mapping percentage can be found in Supplemental Table 1. The sequence files have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE101806.
Statistics.
Statistical analyses were performed using GraphPad Prism 7.0. Comparisons between 2 groups were performed using a 2-tailed unpaired Student’s t test. Comparisons between multiple groups were performed using a 1-way ANOVA with an appropriate post hoc test denoted in the figure legend. To control for multiple comparisons in RNA-Seq data, FDR was calculated. Comparisons were considered significant if P value or FDR q was less than 0.05.
Study approval.
All mouse experiments were performed in accordance with US federal law and approved by the Boston University School of Medicine IACUC (Permit 14859).
Author contributions
GAW, ADS, ACH, NMSS, KLH, ATL, and MRJ conceived and designed the experiments. GAW, ADS, ACH, NMSS, KLH, ATL, and FC performed the experiments. GAW, ADS, ATL, LJQ, JPM, AF, and MRJ analyzed the data. KY, HK, CC, XA, XV, JPM, AF, DO, and MRJ contributed reagents and materials. GAW and MRJ wrote the manuscript with contributions from all authors.
Supplementary Material
Acknowledgments
This work was supported by NIH F31-HL127978 (to GAW), NIH T32-AI089673 (to GAW), NIH R01-HL104053 (to MRJ), NIH R01-HL135756 (to JPM), NIH R01-HL124392 (to XV), NIH R01-HL111449 (to LJQ), and Clinical and Translational Science Institute 1UL1TR001430 (Pilot to MRJ and AF). The research leading to these results has received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement GA 310206. The authors thank Adam Gower for bioinformatics guidance and consultation, Katia Oleinik for assistance in the R environment, and the Boston University and Dana-Farber Cancer Institute Flow Cytometry Core Facilities.
Version 1. 09/14/2017
Electronic publication
Version 2. 09/20/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(10):3866–3876.https://doi.org/10.1172/JCI94639.
Contributor Information
Kazuko Yamamoto, Email: kazukomd@bu.edu.
Hirofumi Kamata, Email: h.kamata142223@gmail.com.
Claudia Carrieri, Email: ccarrier@staffmail.ed.ac.uk.
Xingbin Ai, Email: Xai@partners.org.
Xaralabos Varelas, Email: xvarelas@bu.edu.
Felicia Chen, Email: felichen@bu.edu.
Alan Fine, Email: afine@bu.edu.
References
- 1.Hogan BL, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamata H, et al. Epithelial cell-derived secreted and transmembrane 1a signals to activated neutrophils during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2016;55(3):407–418. doi: 10.1165/rcmb.2015-0261OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 8.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2(6):819–830. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 9.Nolde MJ, Cheng EC, Guo S, Lin H. Piwi genes are dispensable for normal hematopoiesis in mice. PLoS One. 2013;8(8):e71950. doi: 10.1371/journal.pone.0071950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan Y, et al. Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin (Shanghai) 2015;47(5):315–324. doi: 10.1093/abbs/gmv018. [DOI] [PubMed] [Google Scholar]
- 11.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma AK, et al. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97(2):426–434. doi: 10.1182/blood.V97.2.426. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. 2016;196(4):1591–1603. doi: 10.4049/jimmunol.1500805. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs JE, Wagner M, Dhahbi J, Boffelli D, Martin DI. Deficiency of MIWI2 (Piwil4) induces mouse erythroleukemia cell differentiation, but has no effect on hematopoiesis in vivo. PLoS One. 2013;8(12):e82573. doi: 10.1371/journal.pone.0082573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng EC, Kang D, Wang Z, Lin H. PIWI proteins are dispensable for mouse somatic development and reprogramming of fibroblasts into pluripotent stem cells. PLoS One. 2014;9(9):e97821. doi: 10.1371/journal.pone.0097821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto K, et al. Roles of lung epithelium in neutrophil recruitment during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2014;50(2):253–262. doi: 10.1165/rcmb.2013-0114OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasiliauskaitė L, et al. A MILI-independent piRNA biogenesis pathway empowers partial germline reprogramming. Nat Struct Mol Biol. 2017;24(7):604–606. doi: 10.1038/nsmb.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol. 2004;286(4):L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 20.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004;164(2):577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci U S A. 2007;104(2):410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106(31):12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guha A, et al. Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. Proc Natl Acad Sci U S A. 2012;109(31):12592–12597. doi: 10.1073/pnas.1204710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain R, et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrieri C, et al. A transit-amplifying population underpins the efficient regenerative capacity of the testis. J Exp Med. 2017;214(6):1631–1641. doi: 10.1084/jem.20161371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, et al. Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells. 2012;30(9):1948–1960. doi: 10.1002/stem.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- 28.Ma X, et al. Piwi is required in multiple cell types to control germline stem cell lineage development in the Drosophila ovary. PLoS One. 2014;9(3):e90267. doi: 10.1371/journal.pone.0090267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez J, Qi H, Liu N, Lin H. Piwi is a key regulator of both somatic and germline stem cells in the Drosophila testis. Cell Rep. 2015;12(1):150–161. doi: 10.1016/j.celrep.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song H, Yao E, Lin C, Gacayan R, Chen MH, Chuang PT. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A. 2012;109(43):17531–17536. doi: 10.1073/pnas.1207238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5(12):e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy MG, et al. Muc5b is required for airway defence. Nature. 2014;505(7483):412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pittet LA, et al. Earliest innate immune responses require macrophage RelA during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2011;45(3):573–581. doi: 10.1165/rcmb.2010-0210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahy JV, Dickey BF. Airway mucus function and dysfunction. N Engl J Med. 2010;363(23):2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elizur A, Adair-Kirk TL, Kelley DG, Griffin GL, deMello DE, Senior RM. Clara cells impact the pulmonary innate immune response to LPS. Am J Physiol Lung Cell Mol Physiol. 2007;293(2):L383–L392. doi: 10.1152/ajplung.00024.2007. [DOI] [PubMed] [Google Scholar]
- 37.Elizur A, Adair-Kirk TL, Kelley DG, Griffin GL, Demello DE, Senior RM. Tumor necrosis factor-alpha from macrophages enhances LPS-induced clara cell expression of keratinocyte-derived chemokine. Am J Respir Cell Mol Biol. 2008;38(1):8–15. doi: 10.1165/rcmb.2007-0203OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyatt LD, et al. Myeloid ZFP36L1 does not regulate inflammation or host defense in mouse models of acute bacterial infection. PLoS One. 2014;9(10):e109072. doi: 10.1371/journal.pone.0109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto K, et al. Type I alveolar epithelial cells mount innate immune responses during pneumococcal pneumonia. J Immunol. 2012;189(5):2450–2459. doi: 10.4049/jimmunol.1200634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet Journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 41.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.