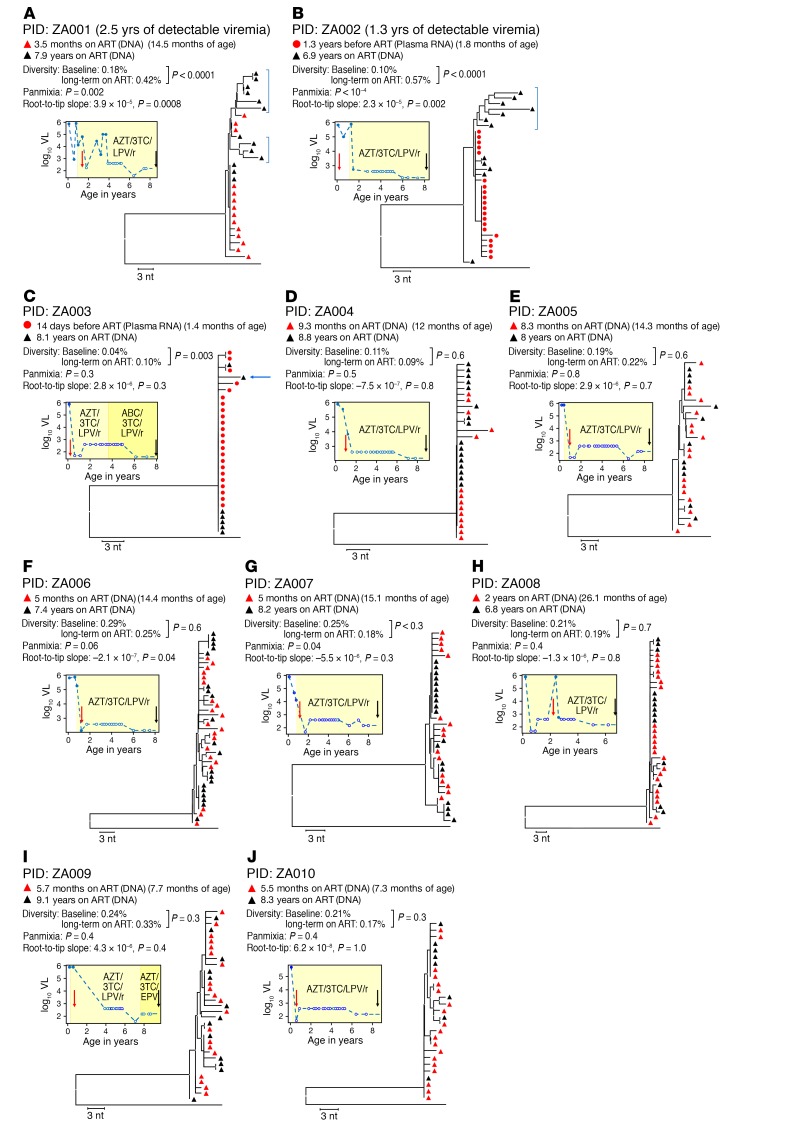
Figure 1. Properties of HIV-1 infection in the 10 children studied.
HIV sequence diversity was measured as the APD at the indicated time points. Divergence was measured using a test for panmixia, with a P value of less than 0.001 considered significant after correcting for multiple comparisons (8), and by measuring the root-to-tip distances of sequences on phylogenetic trees (longer branches over time indicate viral evolution). A 1-sample t test was used to determine if there was a significant change in APD on ART, and a 2-tailed F test was used to determine whether the root-to-tip slopes were significantly different from 0. All branches are shown as the p-distance in the NJ trees. Baseline HIV sequences were either from PBMC DNA (red triangles) or plasma RNA (red circles), and sequences for long-term ART were all from PBMC DNA (black triangles), sampled at the indicated time points. Blue brackets show sequences with evidence of HIV evolution. Plots to the left of the trees show the plasma HIV RNA viral load (VL) as a function of age. Red and black arrows on the plots indicate the time of collection of samples analyzed by SGS. Red vertical arrows indicate the patient’s age at ART initiation. White symbols indicate samples with an undetectable VL, plotted at the limit of sensitivity of the assay used. The apparent decline in the level of undetectable VL in some samples is due to a switch to an assay with a lower limit of detection. (A and B) Patients with detectable viremia for more than 1 year after the time of the first sample. (C–J) Patients with undetectable viremia for all samples taken after the first sample. The blue arrow indicates a sequence that was omitted in 1 sensitivity analysis, as discussed in the text. Results from a total of 20 samples are included in this figure.