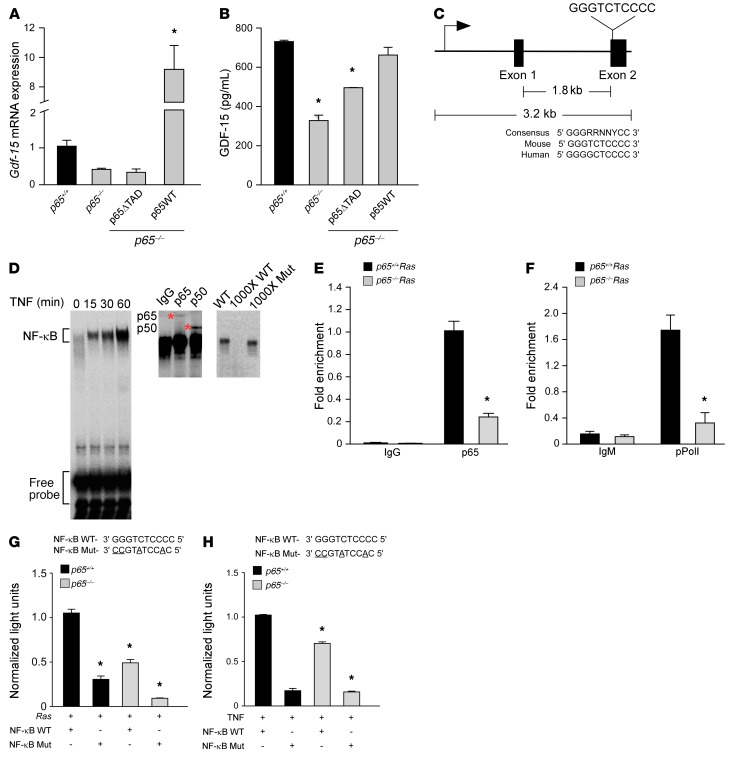
Figure 4. NF-κB is a direct regulator of Gdf-15.
(A) p65–/– MEFs were infected with retrovirus for full-length (p65WT) or truncated p65 (p65ΔTAD). Gdf-15 expression by qRT-PCR was assayed following TNF treatment (5 ng/ml) for 2 hours. n = 3. Data are shown as mean ± SEM. *P ≤ 0.005, 1-way ANOVA. (B) ELISA from conditioned media from transfected cells in A. n = 3. Data are shown as mean ± SEM. *P ≤ 0.05, 1-way ANOVA. (C) Schematic of Gdf-15 gene with NF-κB consensus site in exon 2, compared with consensus site in mouse and human. (D) EMSA from TNF-treated Ras MEFs using NF-κB consensus site probe shown in C. Supershift assay from Ras MEFs incubated with antisera specific for p65 and p50. Asterisks indicate supershifted complexes. Specificity of the complexes was tested by adding ×1000 molar excess of labeled WT probe or nonlabeled mutant probe. (E) ChIP assays for p65 binding from Ras MEFs. DNA was amplified with oligonucleotides spanning the NF-κB site on exon 2 of Gdf-15. Fold enrichment over IgG controls (normalized to input) are indicated. n = 3. *P ≤ 0.05, Student’s t test. (F) ChIP for pPol II (serine 2 version), as described in Figure 4E. DNA was amplified with the same oligonucleotides as shown in E. Fold enrichment over IgM controls (normalized to input) are indicated. n = 3. *P ≤ 0.05. Student’s t test. (G) MEFs were transfected with a luciferase reporter with WT or mutated NF-κB consensus sites, as shown in C. Cells were cotransfected with H-RasG12V, and after 48 hours, luciferase activity was measured. n = 3. *P ≤ 0.05 compared with p65+/+ MEFs with WT construct+ H-Ras, 2-way ANOVA. (H) MEFs were transfected with WT and mutant luciferase reporters and treated with 1 μl/ml of TNF for 2 hours. Luciferase activity was measured. n = 3. *P ≤ 0.05 when compared with p65+/+ MEFs with WT construct with TNF, 2-way ANOVA.