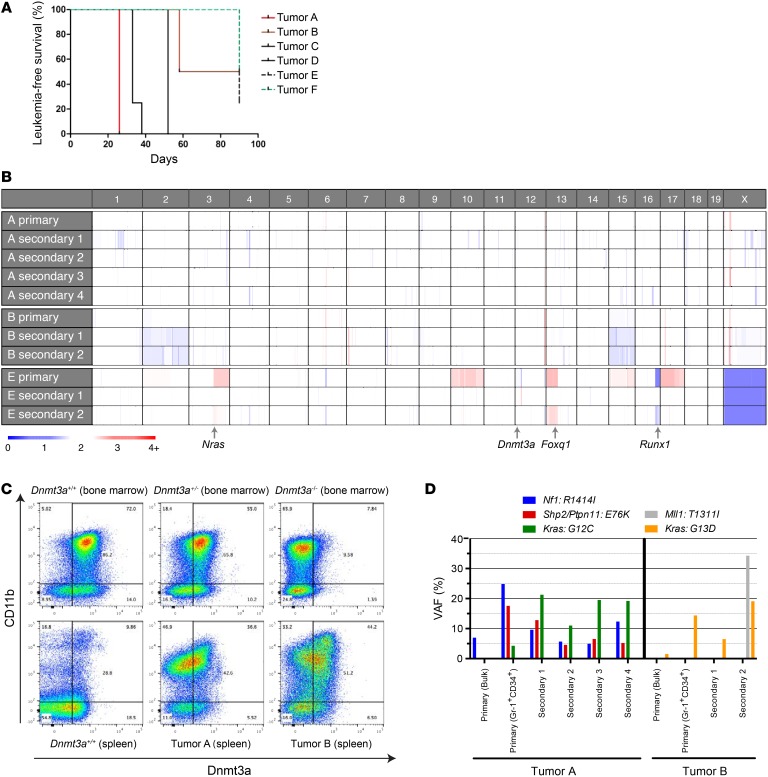
Figure 6. Persistent expression of the residual WT Dnmt3a gene in AML samples arising in Dnmt3a+/– mice.
(A) Plot demonstrating disease latency for sublethally irradiated WT animals engrafted with Dnmt3a+/– tumors, designated A–F (see Table 1 and Supplemental Table 2). For each of the 6 primary tumors that were transplanted, 3 to 5 secondary recipient mice were assessed. (B) Copy number variation in sequenced tumors. Note that none of the tumors has deletions involving chromosome 12 at the location of the Dnmt3a gene. The locations of 3 cancer-related genes that were amplified (Nras, Foxq1) or deleted (Runx1) in tumor E are shown. Tumor E was derived from a male mouse; the single copy of the X chromosome in these tumors “calibrates” the color value for a single copy deletion. (C) Representative flow cytometry plots for Dnmt3a protein abundance in the CD11b+ compartment of Dnmt3a+/+, Dnmt3a+/–, and Dnmt3a–/– bone marrow samples (top panels) and a WT spleen or Dnmt3a+/– tumors A and B (derived from the unmanipulated spleen samples from the primary mice), showing preserved expression of WT Dnmt3a protein in the CD11b+ cells in each tumor spleen sample. The level of Dnmt3a protein in the tumor cells was similar to that of the haploinsufficient bone marrow cells. (D) VAFs for selected mutations detected in primary tumors (either bulk or sorted to enrich for Gr-1+CD34+ myeloid tumor cells) and in corresponding tumors from transplanted secondary recipients. Kras mutation VAFs are from AmpliSeq data (Supplemental Table 6), while other mutation VAFs are from exome sequencing (Supplemental Tables 3–5).