Bacteria are prolific producers of secondary metabolites, a group of molecules that have captivated chemists and biologists alike with their fascinating structures, unusual biosynthetic pathways, and potent biological activities. Secondary metabolites, also termed natural products, are produced by dedicated biosynthetic gene clusters (BGCs), and the recent explosion in microbial genome sequences shows that the biosynthetic potential of bacteria has been vastly underestimated. We now know that across microbial genomes only a small fraction of BGCs are actively expressed, while the majority, known as “silent” or “cryptic”, do not give rise to appreciable levels of a small molecule product.1 This leads to two profound conclusions. (1) Though secondary metabolites have been a major source of drug leads, their discovery from microbial sources has inadvertently focused on a small subset of constitutively active BGCs. (2) The chemistry and biology of most BGCs remain largely unexplored. Understanding how these gene clusters are regulated and what cues activate them could provide new insights into microbial physiology and significantly impact natural products research and, by extension, drug discovery. However, strategies that can address these multifaceted aspects of cryptic BGCs are only starting to emerge.
To investigate these unresolved issues underlying cryptic BGCs, we recently extended a method known as HiTES (high-throughput elicitor screens) to Streptomyces spp., a genus known for its rich biosynthetic potential.2,3 Previous strategies for awakening silent clusters have predominantly centered on finding the natural products that they encode, usually via heterologous expression or insertion of exogenous promoter elements. In contrast, HiTES uses endogenous regulatory pathways for inducing silent gene clusters. As such, it has the ability to pinpoint both the signals that activate a given BGC and the resulting small molecule product. In this method, a genetic reporter is placed inside the gene cluster of interest, thus providing a rapid readout for its level of expression. The reporter strain is then screened against a library of small molecules in an effort to identify elicitors. With such elicitors in hand, the cryptic metabolite and the mechanism of induction can be identified.2
To implement HiTES in streptomycetes, we chose a cryptic nonribosomal peptide synthetase BGC (sur) in Streptomyces albus.3 Because many streptomycetes naturally encode β-galactosidases, an enhanced green fluorescent protein (eGFP) reporter was used, rather than lacZ as in previous iterations of HiTES. Two versions of this eGFP reporter were constructed and screened against a library of roughly 500 natural products or derivatives thereof. Several elicitors, including the anticancer drug etoposide and the antiparasitic agent ivermectin, were identified and subsequently verified using quantitative reverse transcription polymerase chain reaction experiments, which showed upregulation of sur in response to both cytotoxins. We then examined the products of the sur cluster and the mechanisms that lead to its activation. Eighteen cryptic metabolites were elucidated using a combination of nuclear magnetic resonance spectroscopy and mass spectrometry. They fall into five groups: the cyclic surugamides, the acyl-surugamides, the linear surugamide F variants, the albucylones, and albuquinone/mansouramycin analogues. Of these, 14 are novel. The diversity of compounds observed highlights the potential of the sur BGC to amplify its biosynthetic output via crosstalk with other BGCs, implying that ivermectin and etoposide globally activate secondary metabolism in S. albus.3
In addition to providing chemical novelty, several of the cryptic metabolites also exhibited pronounced biological activities, with the cyclic surugamides inhibiting cathepsin B, a protease implicated in cancer, and acyl-surugamide A acting as a potent antifungal agent. Perhaps most interestingly, we have begun to illuminate the regulatory pathways that connect the provision of old natural products, etoposide and ivermectin, with the production of new, cryptic ones. A pathway-specific transcriptional regulator, surR, was found to act as a repressor of the sur BGC; both etoposide and ivermectin downregulate the expression of surR through a mechanism that has yet to be determined. Preliminary evidence also suggests that the SOS response, a stress response triggered upon detection of DNA damage, may be involved in the regulatory circuits that lead to the products of the silent sur cluster.3
These findings share several common features with our previous work in the Gram-negative model bacterium Burkholderia thailandensis. In both cases, low levels of cytotoxins were found to act as pleiotropic inducers of silent BGCs, pointing perhaps to a common theme of antibiotics inducing antibiotics (Figure 1). These results carry obvious implications for the natural roles of antibiotics, a matter that is still under debate.4 Moreover, the pleiotropic nature of the elicitors underlines the poly-pharmacology of these cytotoxins and implicates one or more fundamental regulatory pathways that can globally enhance secondary metabolism. For example, induction of the SOS response by subinhibitory concentrations of antibiotics, which may then induce silent BGCs, could represent one such pathway that can regulate gene expression at a cellwide level. While intriguing, the connection among antibiotics, stress responses, and cryptic metabolites awaits validation and will undoubtedly be the subject of future research (Figure 1).
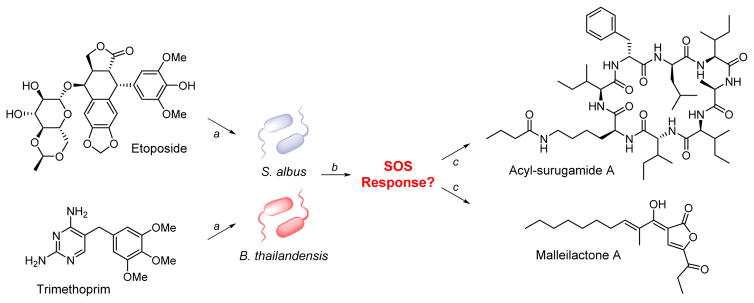
Figure 1.
Activation of silent BGCs by subinhibitory concentrations of antibiotics. (a) Using HiTES, the antibiotics etoposide and trimethoprim were found to be inducers of cryptic BGCs in S. albus and B. thailandensis, respectively. (b) Both antibiotics globally upregulate secondary metabolism, perhaps via the SOS response. (c) Activation of silent gene clusters leads to biosynthesis of the cryptic antifungal acyl-surugamide A (in S. albus) and the antiparasitic agent malleilactone A (in B. thailandensis), among other metabolites.
In a general sense, the interplay between natural products and microbes is poorly understood.4,5 Although the effects of synthetic and natural small molecules on signaling pathways in eukaryotes are the focus of tremendous attention, the effects of these same molecules on bacteria or bacterial communities have remained relatively obscure. This disparity has perhaps been driven by the frequent use of diversity-oriented synthetic small molecule libraries in interrogating eukaryotic signaling pathways relevant in cancer and other disorders. But with recent insights into the importance of microbial assemblies in human health and disease, chemical microbiology—the assessment of how small molecules perturb signaling pathways in bacteria—will come into full swing. As shown by Xu et al.,3 the less-heralded signaling properties of known, therapeutic molecules may be leveraged to activate secondary metabolite production in streptomycetes, which contain a treasure trove of silent gene clusters. Expanding the application of HiTES and deciphering the biological mechanisms that regulate cryptic BGCs may enable the discovery of new, potentially useful drug leads and simultaneously provide insights into the small molecule exchanges that govern the microbial world.
Acknowledgments
Funding
M.R.S. thanks the National Institutes of Health (Grant DP2-AI-124786), the Searle Scholars Program, and the Princeton University IP Accelerator Fund for generous support of this work.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J Am Chem Soc. 2017;139:9203–9212. doi: 10.1021/jacs.7b02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 5.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]